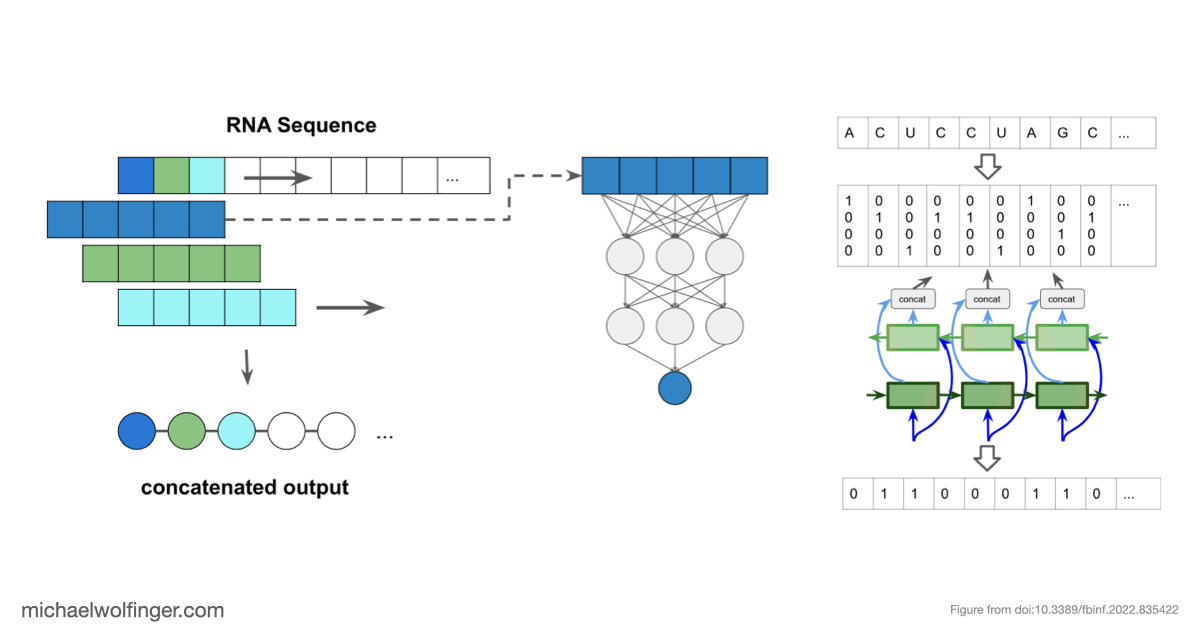
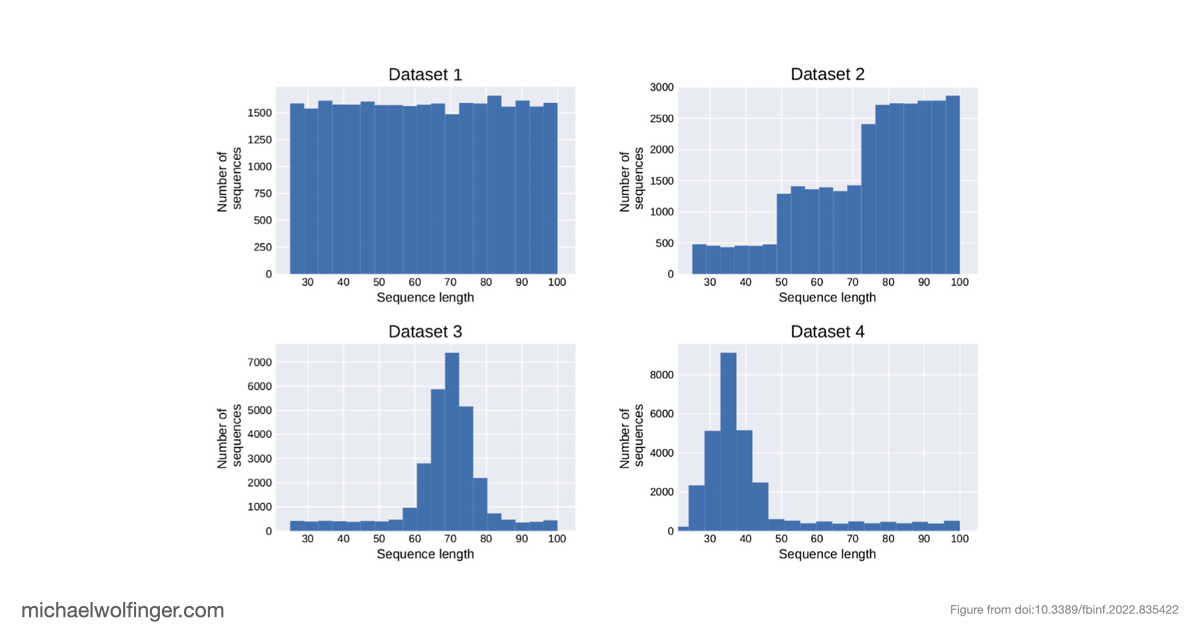
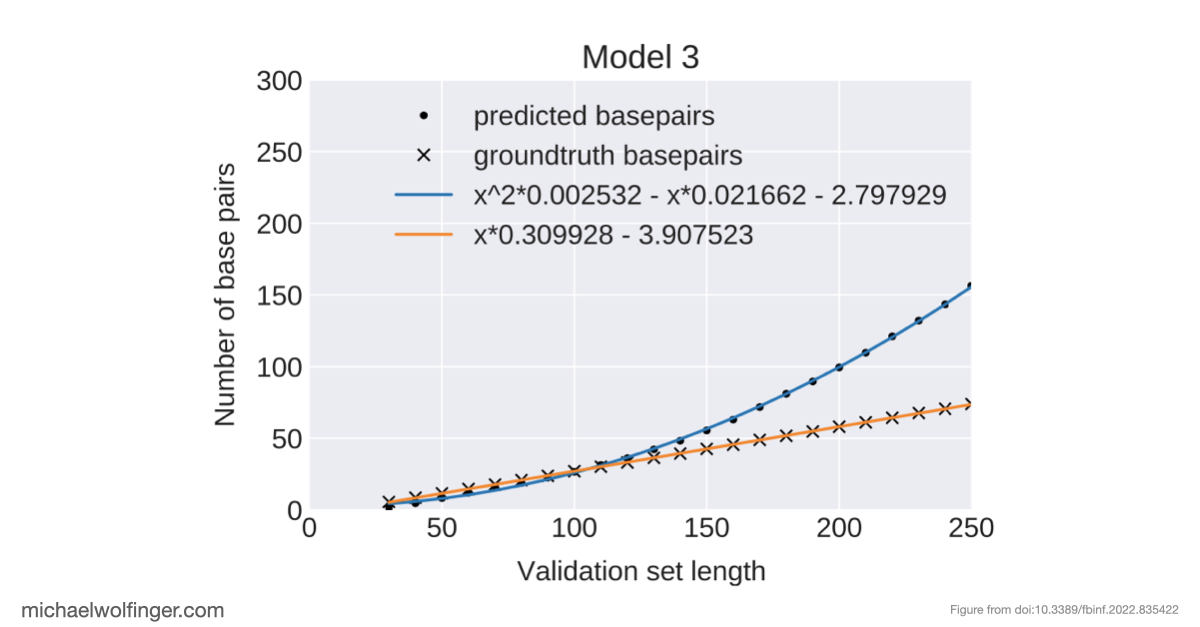
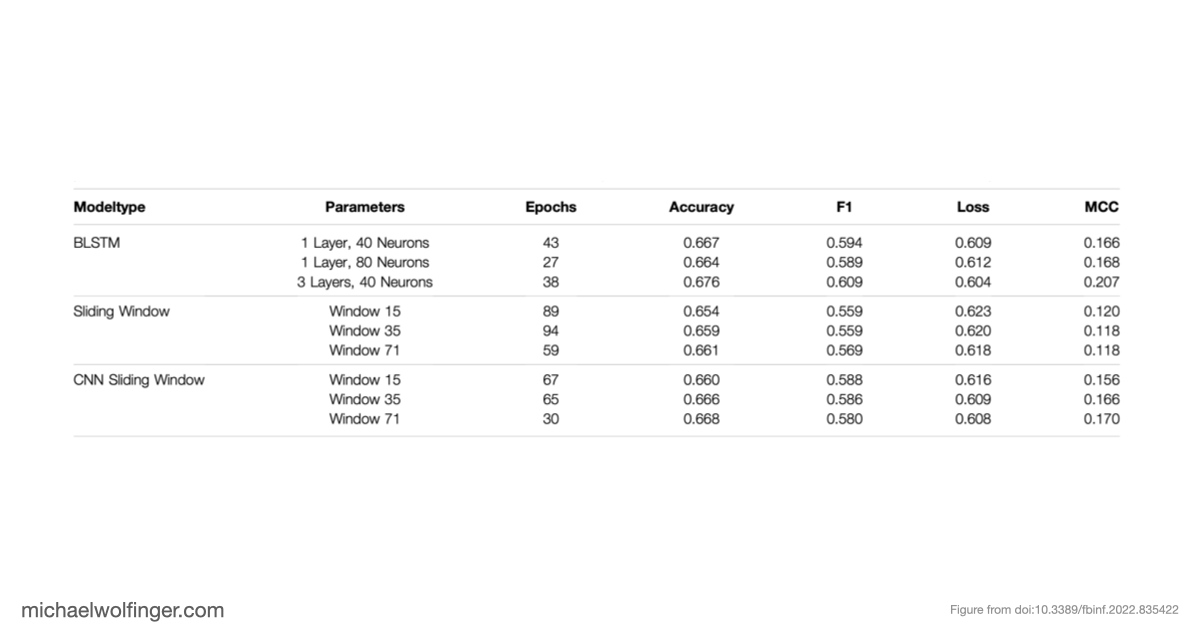
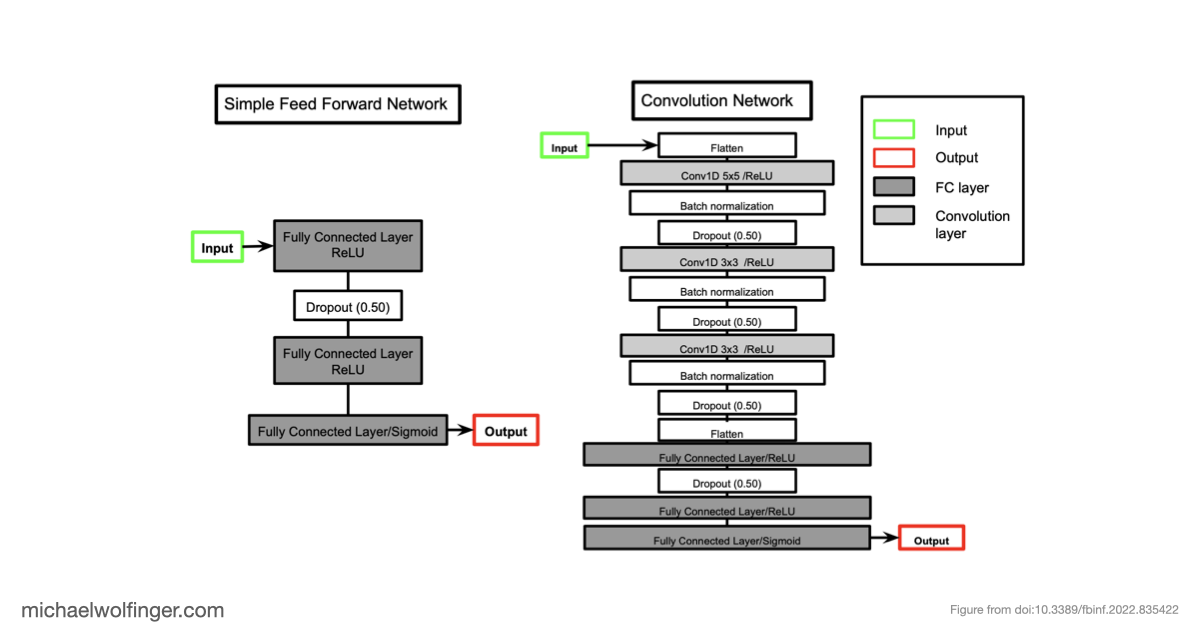
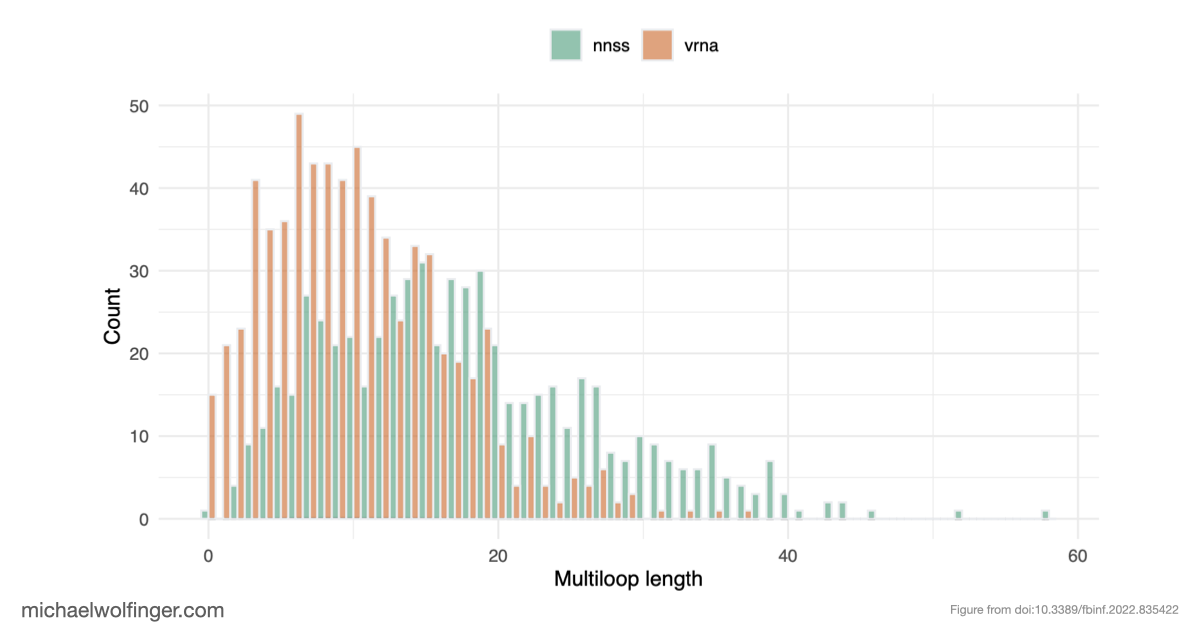
Deep learning methods are unable to predict RNA secondary structures
Machine learning of RNA structure is more challenging than you might think. Using synthetic data from ViennaRNA's RNAfold to study the capabilities and shortcomings of neural networks for RNA secondary structure prediction in a controlled setting, we argue that shortcomings in the artificial setting will translate to real data
Published AI approaches for RNA structure prediction suffer from massively biased training sets, resulting in severely degraded prediction quality on arbitrary RNAs. Using inverse RNA folding, i.e. generating sequences that are compatible with a given structure, we generate synthetic data with the same bias as published deep learning approaches. A network trained on this set performs well on sequences that have no sequence similarity but fold into structures contained in the training set. On sequences with unrelated structures performance falls drastically. Thus, the network generalizes well to new sequences, but not to new structures.
Intriguingly, published deep learning methods on unbiased sets are not even capable of predicting correct RNA base pairing, a problem that is much simpler than the RNA folding problem. On top of that, BLSTMs predict pseudoknots and base triplets, although they do not occur in the ViennaRNA RNAfold ground truth.
Read the full story in our article 'Caveats to Deep Learning Approaches to RNA Secondary Structure Prediction', published in 'Frontiers in Bioinformatics'.
Figures and Data
Citation
Caveats to deep learning approaches to RNA secondary structure prediction
Christoph Flamm, Julia Wielach, Michael T. Wolfinger, Stefan Badelt, Ronny Lorenz, Ivo L. Hofacker
Front. Bioinform. 2:835422 (2022) | doi:10.3389/fbinf.2022.835422 | PDF