Publications by Michael T. Wolfinger
Submitted Manuscripts / Preprints
57. Identification of a region within the 5’UTR of of Interleukin–2 mRNA offers a promising strategy for drug development for autoimmune disorders
Katarzyna M. Gluchowska, Krzysztofa Odzywol, Agnieszka Zagozdzon, Tomasz K. Wirecki, Chandran Nithin, Bartlomiej Surpeta, Xiaobing Zhang, Jannan Zhao, Lukasz Joachimiak, Bartlomiej Hofman, Joanna Sztuba-Solinska, Katarzyna Drzewicka, Angelika Muchowicz, Michael T. Wolfinger, Roman Blaszczyk, Irina Tuszynska, Janusz M. Bujnicki, and Zbigniew Zaslona
Submitted manuscript (2026)
56. Rational design of mechanically active RNAs: de novo engineering of functional exoribonuclease-resistant RNAs
Jule Walter, Leonhard Sidl, Katrin Gutenbrunner, Denis Skibinski, Tim Kolberg, Ivo L. Hofacker, Hua-Ting Yao, Mario Mörl, Michael T. Wolfinger
bioRxiv (2026)
doi:10.64898/2026.01.08.698366 | Preprint PDF
55. “Circle ofLife” - Zika virus genomic cyclization is controlled by sequence specificity
Liam Kerr, Danielle L. Gemmill, Higor S. Pereira, Michael T. Wolfinger, Trushar R. Patel
Submitted manuscript (2025)
- - -
Peer-reviewed journal / conference articles and book chapters
2025
54. Identification of conserved RNA regulatory switches in living cells using RNA secondary structure ensemble mapping and covariation analysis
Ivana Borovská, Chundan Zhang, Sarah-Luisa J. Dülk, Edoardo Morandi, Marta F. S. Cardoso, Billal M. Bourkia, Daphne A. L. van den Homberg, Michael T. Wolfinger, Willem A. Velema, Danny Incarnato
Nat. Biotechnol. (2025)
doi:10.1038/s41587-025-02739-0 | PDF | Journal article
53. From Structure to Function: Computational Insights into Musashi-RNA Complexes in the Context of Viral Pathogenesis and Beyond
Nitchakan Darai, Leonhard Sidl, Thanyada Rungrotmongkol, Peter Wolschann, Michael T. Wolfinger
Sci. Asia 51S(1) 2025s013:1-10 (2025)
doi:10.2306/scienceasia1513-1874.2025.s013 | PDF | Review article
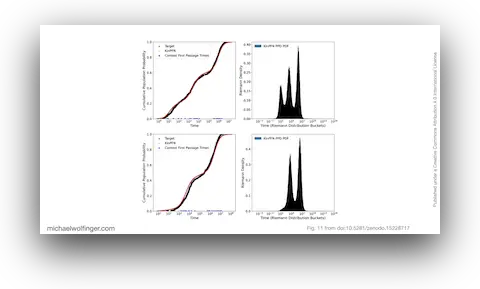
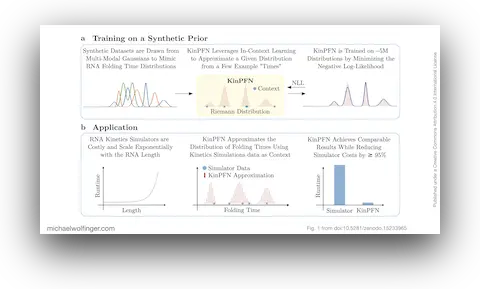
52. Bayesian Approximation of RNA Folding Times
Dominik Scheuer, Frederic Runge, Jörg K.H. Franke, Michael T. Wolfinger, Christoph Flamm, Frank Hutter
ICLR 2025 Workshop on AI for Nucleic Acids (2025)
doi:10.5281/zenodo.15228717 | PDF | Conference article
51. KinPFN: Bayesian Approximation of RNA Folding Kinetics using Prior-Data Fitted Networks
Dominik Scheuer, Frederic Runge, Jörg K.H. Franke, Michael T. Wolfinger, Christoph Flamm, Frank Hutter
The Thirteenth International Conference on Learning Representations (ICLR'25) (2025)
doi:10.5281/zenodo.15233965 | PDF | Conference article
2024
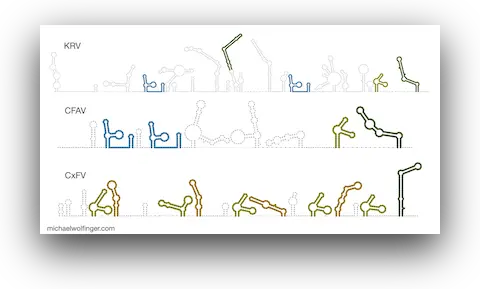
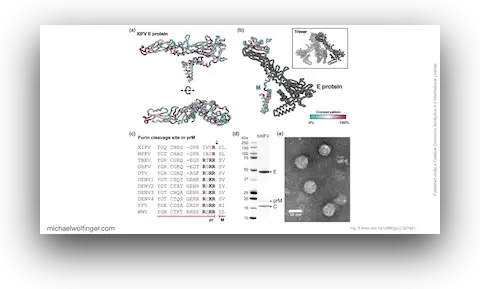
50. Pan-flavivirus analysis reveals sfRNA-independent, 3’UTR-biased siRNA production from an Insect-Specific Flavivirus
Benoit Besson, Gijs J. Overheul, Michael T. Wolfinger, Sandra Junglen, Ronald P. van Rij
J. Virol. e01215-24 (2024)
doi:10.1128/jvi.01215-24 | Preprint PDF | Journal article
49. Xinyang flavivirus, from Haemaphysalis flava ticks in Henan province, China, defines a basal, likely tick-only flavivirus clade
Lan-Lan Wang, Qia Cheng, Natalee D. Newton, Michael T. Wolfinger, Mahali S. Morgan, Andrii Slonchak, Alexander A. Khromykh, Tian-Yin Cheng, Rhys H. Parry
J. Gen. Virol. 105(5) (2024)
doi:10.1099/jgv.0.001991 | PDF | Journal article
48. A framework for automated scalable designation of viral pathogen lineages from genomic data
Jakob McBroome, Adriano de Bernardi Schneider, Cornelius Roemer, Michael T. Wolfinger, Angie S. Hinrichs, Aine N. O’Toole, Chris Ruis, Yatish Turakhia, Andrew Rambaut, and Russell Corbett-Detig
Nature Microbiol. 9:550–560 (2024)
doi:10.1038/s41564-023-01587-5 | PDF | Journal article
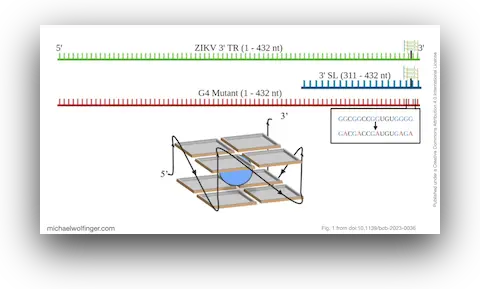
47. The 3’ terminal region of Zika virus RNA contains a conserved G-quadruplex and is unfolded by human DDX17
Danielle L. Gemmill, Corey R. Nelson, Maulik D. Badmalia, Higor S. Pereira, Michael T. Wolfinger, and Trushar Patel
Biochem. Cell Biol. 102(1):96–105 (2024)
doi:10.1139/bcb-2023-0036 | PDF | Journal article
2023
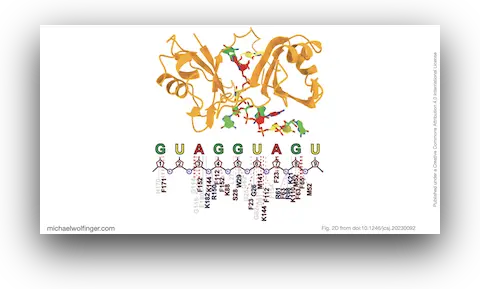
46. A Structural Refinement Technique for Protein-RNA Complexes Using a Combination of AI-based Modeling and Flexible Docking: A Study of Musashi-1 Protein
Nitchakan Darai, Kowit Hengphasatporn, Peter Wolschann, Michael T. Wolfinger, Yasuteru Shigeta, Thanyada Rungrotmongkol, Ryuhei Harada
B. Chem. Soc. Jpn. 96(7):677–685 (2023)
doi:10.1246/bcsj.20230092 | PDF | Journal article
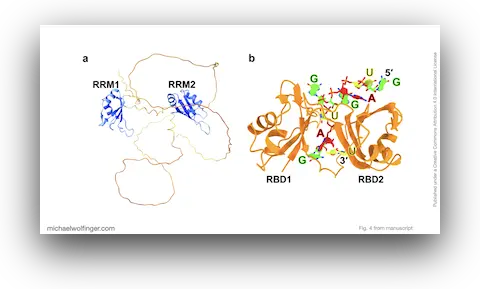
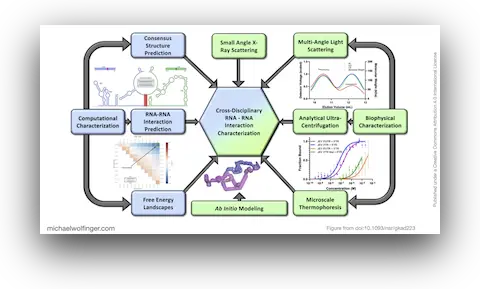
45. Investigating RNA-RNA interactions through computational and biophysical analysis
Tyler Mrozowich, Sean Park, Maria Waldl, Amy Henrickson, Scott Tersteeg, Corey R. Nelson, Anneke De Klerk, Borries Demeler, Ivo L. Hofacker, Michael T. Wolfinger, Trushar R. Patel
Nucleic Acids Res. 51(9):4588–4601 (2023)
doi:10.1093/nar/gkad223 | PDF | Supplement | Figures | Journal article
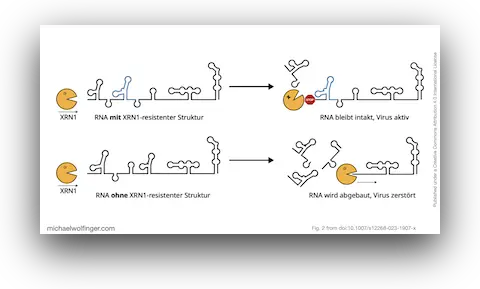
44. Strukturierte RNAs in Viren (in German)
Roman Ochsenreiter, Michael T. Wolfinger
Biospektrum 29(2):156-158 (2023)
doi:10.1007/s12268-023-1907-x | PDF | Figures | Review article
2022
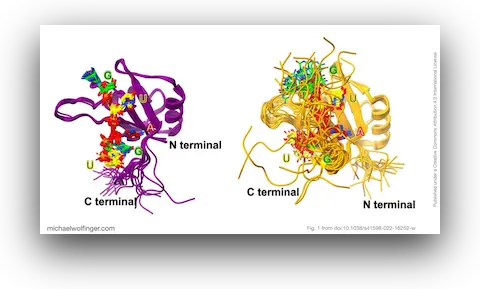
43. Theoretical studies on RNA recognition by Musashi 1 RNA–binding protein
Nitchakan Darai, Panupong Mahalapbutr, Peter Wolschann, Vannajan Sanghiran Lee, Michael T. Wolfinger, Thanyada Rungrotmongkol
Sci. Rep. 12:12137 (2022)
doi:10.1038/s41598-022-16252-w | PDF | Figures | Journal article
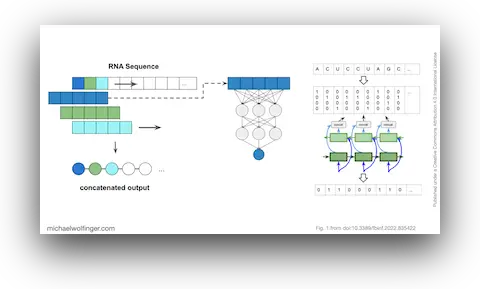
42. Caveats to deep learning approaches to RNA secondary structure prediction
Christoph Flamm, Julia Wielach, Michael T. Wolfinger, Stefan Badelt, Ronny Lorenz, Ivo L. Hofacker
Front. Bioinform. 2:835422 (2022)
doi:10.3389/fbinf.2022.835422 | PDF | Figures | Journal article
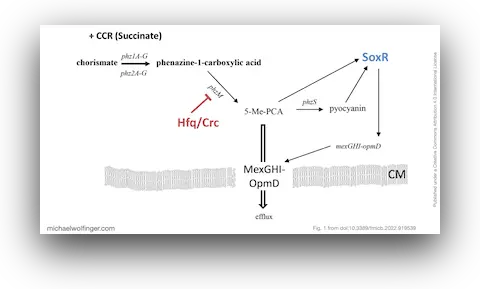
41. Rewiring of Gene Expression in Pseudomonas aeruginosa During Diauxic Growth Reveals an Indirect Regulation of the MexGHI-OpmD Efflux Pump by Hfq
Marlena Rozner, Ella Nukarinen, Michael T. Wolfinger, Fabian Amman, Wolfram Weckwerth, Udo Blaesi, Elisabeth Sonnleitner
Front. Microbiol. 13:919539 (2022)
doi:10.3389/fmicb.2022.919539 | PDF | Journal article
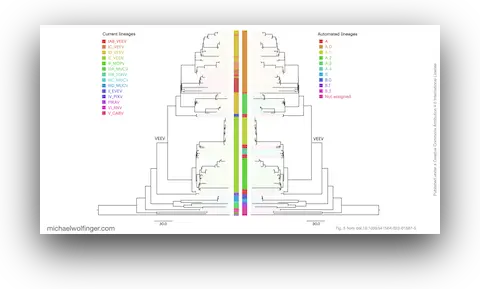
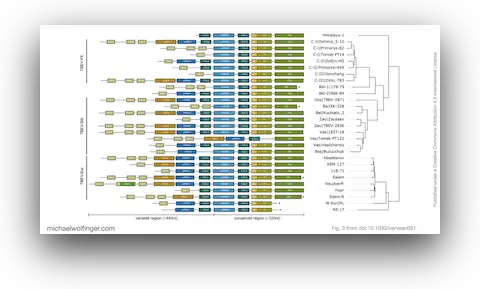
40. Evolutionary traits of Tick-borne encephalitis virus: Pervasive non-coding RNA structure conservation and molecular epidemiology
Lena S. Kutschera, Michael T. Wolfinger
Virus Evol. (8):1 veac051 (2022)
doi:10.1093/ve/veac051 | PDF | Figures | Journal article
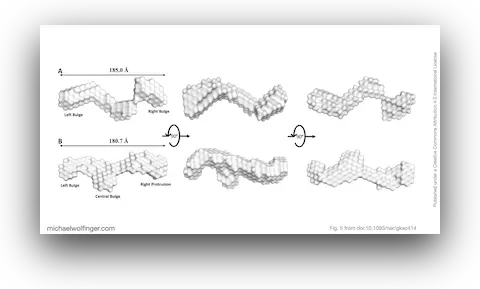
39. Biophysical Characterisation of Human LincRNA-p21 Sense and Antisense Alu Inverted Repeats
Michael H. D’Souza, Tyler Mrozowich, Maulik D. Badmalia, Mitchell Geeraert, Angela Frederickson, Amy Henrickson, Borries Demeler, Michael T. Wolfinger, and Trushar R. Patel
Nucleic Acids Res. 50(10):5881–5898 (2022)
doi:10.1093/nar/gkac414 | PDF | Journal article
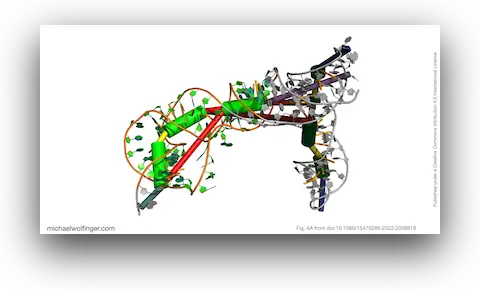
38. Insights into the secondary and tertiary structure of the Bovine Viral Diarrhea Virus Internal Ribosome Entry Site
Devadatta Gosavi, Iwona Wower, Irene K. Beckmann, Ivo L. Hofacker, Jacek Wower, Michael T. Wolfinger, Joanna Sztuba-Solinska
RNA Biol. 19(1) 496-506 (2022)
doi:10.1080/15476286.2022.2058818 | PDF | Figures | Journal article
2021
37. Functional RNA Structures in the 3’UTR of Mosquito-Borne Flaviviruses
Michael T. Wolfinger, Roman Ochsenreiter, Ivo L. Hofacker
In Virus Bioinformatics, edited by Dmitrij Frishman and Manja Marz, pp65–100. Chapman and Hall/CRC Press (2021)
doi:10.1201/9781003097679-5 | Preprint PDF | Figures | Book chapter
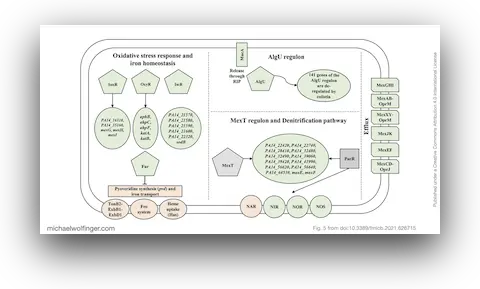
36. Gene Expression Profiling of Pseudomonas Aeruginosa Upon Exposure to Colistin and Tobramycin
Anastasia Cianciulli Sesso, Branislav Lilić, Fabian Amman, Michael T. Wolfinger, Elisabeth Sonnleitner, Udo Bläsi
Front. Microbiol. 12:937 (2021)
doi:10.3389/fmicb.2021.626715 | PDF | Journal article
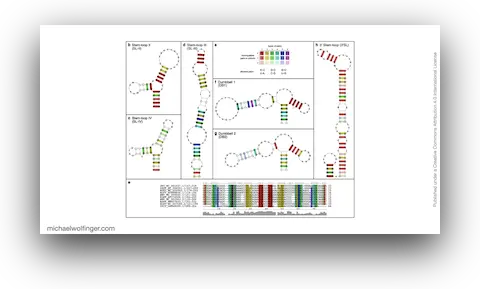
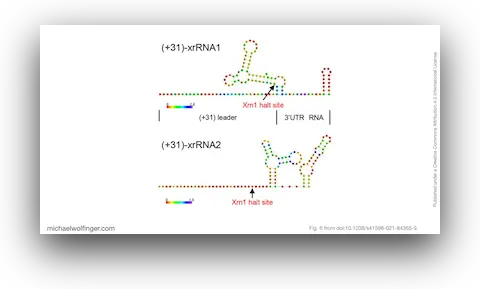
35. An African Tick Flavivirus Forming an Independent Clade Exhibits Unique Exoribonuclease-Resistant RNA Structures in the Genomic 3’-Untranslated Region
Hayato Harima, Yasuko Orba, Shiho Torii, Yongjin Qiu, Masahiro Kajihara, Yoshiki Eto, Naoya Matsuta, Bernard M. Hang’ombe, Yuki Eshita, Kentaro Uemura, Keita Matsuno, Michihito Sasaki, Kentaro Yoshii, Ryo Nakao, William W. Hall, Ayato Takada, Takashi Abe, Michael T. Wolfinger, Martin Simuunza, Hirofumi Sawa
Sci. Rep. 11:4883 (2021)
doi: 10.1038/s41598-021-84365-9 | PDF | Journal article
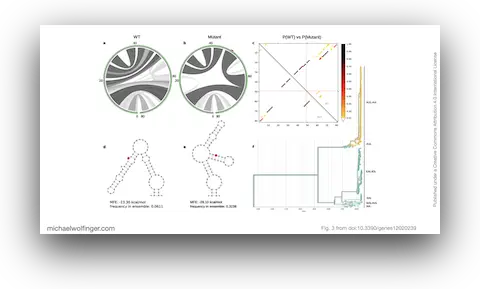
34. Dynamic Molecular Epidemiology Reveals Lineage-Associated Single-Nucleotide Variants That Alter RNA Structure in Chikungunya Virus
Thomas Spicher, Markus Delitz, Adriano de Bernardi Schneider, Michael T. Wolfinger
Genes 12 (2):239 (2021)
doi:10.3390/genes12020239 | PDF | Figures | Journal article
2020
33. Bi-Alignments as Models of Incongruent Evolution of RNA Sequence and Secondary Structure
Maria Waldl, Sebastian Will, Michael T. Wolfinger, Ivo L. Hofacker, Peter F. Stadler
In Computational Intelligence Methods for Bioinformatics and Biostatistics, pp159–70. Springer International Publishing (2020)
doi:10.1007/978-3-030-63061-4_15 | Preprint PDF | Conference article
32. Genomic Epidemiology of Superspreading Events in Austria Reveals Mutational Dynamics and Transmission Properties of SARS-CoV-2
Alexandra Popa, Jakob-Wendelin Genger, Michael D. Nicholson, Thomas Penz, Daniela Schmid, Stephan W Aberle, Benedikt Agerer, Alexander Lercher, Lukas Endler, Henrique Colaco, Mark Smyth, Michael Schuster, Miguel L. Grau, Francisco Martínez-Jiménez, Oriol Pich, Wegene Borena, Erich Pawelka, Zsofia Keszei, Martin Senekowitsch, Jan Laine, Judith H Aberle, Monika Redlberger-Fritz, Mario Karolyi, Alexander Zoufaly, Sabine Maritschnik, Martin Borkovec, Peter Hufnagl, Manfred Nairz, Günter Weiss, Michael T. Wolfinger, Dorothee von Laer, Giulio Superti-Furga, Nuria Lopez-Bigas, Elisabeth Puchhammer-Stöckl, Franz Allerberger, Franziska Michor, Christoph Bock, Andreas Bergthaler
Sci. Transl. Med. 12 (573):eabe2555 (2020)
doi:10.1126/scitranslmed.abe2555 | PDF | Journal article
31. Discoveries of Exoribonuclease-Resistant Structures of Insect-Specific Flaviviruses Isolated in Zambia
Christida E. Wastika, Hayato Harima, Michihito Sasakai, Bernard M. Hang’ombe, Yuki Eshita, Qiu Yongjin, William W. Hall, Michael T. Wolfinger, Hirofumi Sawa, Yasuko Orba
Viruses 12:1017 (2020)
doi:10.3390/v12091017 | PDF | Journal article
30. Distinctive Regulation of Carbapenem Susceptibility in Pseudomonas Aeruginosa by Hfq
Elisabeth Sonnleitner, Petra Pusic, Michael T. Wolfinger, Udo Bläsi
Front. Microbiol. 11:1001 (2020)
doi:10.3389/fmicb.2020.01001 | PDF | Journal article
2019
29. Updated Phylogeny of Chikungunya Virus Suggests Lineage-Specific RNA Architecture
Adriano de Bernardi Schneider, Roman Ochsenreiter, Reilly Hostager, Ivo L. Hofacker, Daniel Janies, Michael T. Wolfinger
Viruses 11:798 (2019)
doi:10.3390/v11090798 | PDF | Figures | Journal article
28. Musashi Binding Elements in Zika and Related Flavivirus 3’UTRs: A Comparative Study in Silico
Adriano de Bernardi Schneider, Michael T. Wolfinger
Sci. Rep. 9(1):6911 (2019)
doi:10.1038/s41598-019-43390-5 | PDF | Figures | Journal article
27. Indications for a Moonlighting Function of Translation Factor aIF5A in the Crenarchaeum Sulfolobus Solfataricus
Flavia Bassani, Isabelle Anna Zink, Thomas Pribasnig, Michael T. Wolfinger, Alice Romagnoli, Armin Resch, Christa Schleper, Udo Bläsi, Anna La Teana
RNA Biol. 16 (5):675–85 (2019)
doi:10.1080/15476286.2019.1582953 | PDF | Journal article
26. Functional RNA Structures in the 3’UTR of Tick-Borne, Insect-Specific and No Known Vector Flaviviruses
Roman Ochsenreiter, Ivo L. Hofacker, Michael T. Wolfinger
Viruses 11:298 (2019)
doi:10.3390/v11030298 | PDF | Figures | Journal article
2018
25. Harnessing Metabolic Regulation to Increase Hfq-Dependent Antibiotic Susceptibility in Pseudomonas Aeruginosa
Petra Pusic, Elisabeth Sonnleitner, Beatrice Krennmayr, Dorothea Agnes Heitzinger, Michael T. Wolfinger, Armin Resch, Udo Bläsi
Front. Microbiol. 9:2709 (2018)
doi:10.3389/fmicb.2018.02709 | PDF | Journal article
24. TERribly Difficult: Searching for Telomerase RNAs in Saccharomycetes
Maria Waldl, Bernhard C. Thiel, Roman Ochsenreiter, Alexander Holzenleiter, João Victor de Araujo Oliveira, Maria Emília M.T. Walter, Michael T. Wolfinger, Peter F. Stadler
Genes 9 (8), 372 (2018)
doi:10.3390/genes9080372 | PDF | Journal article
23. Efficient Computation of Cotranscriptional RNA-Ligand Interaction Dynamics
Michael T. Wolfinger, Christoph Flamm, Ivo L. Hofacker
Methods 143:70–76 (2018)
doi:10.1016/j.ymeth.2018.04.036 | Preprint PDF | Journal article
22. In Silico Design of Ligand Triggered RNA Switches
Sven Findeiß, Stefan Hammer, Michael T. Wolfinger, Felix Kühnl, Christoph Flamm, Ivo L. Hofacker
Methods 143:90–101 (2018)
doi:10.1016/j.ymeth.2018.04.003 | Preprint PDF | Journal article
21. Interplay Between the Catabolite Repression Control Protein Crc, Hfq and RNA in Hfq-Dependent Translational Regulation in Pseudomonas Aeruginosa
Elisabeth Sonnleitner, Alexander Wulf, Sébastien Campagne, Xue-Yuan Pei, Michael T. Wolfinger, Giada Forlani, Konstantin Prindl, Laetitia Abdou, Armin Resch, Frederic Allain, Ben Luisi, Henning Urlaub, Udo Bläsi
Nucleic Acids Res. 46:1470–85 (2018)
doi:10.1093/nar/gkx1245 | PDF | Journal article
2017
20. The Anaerobically Induced sRNA PaiI Affects Denitrification in Pseudomonas Aeruginosa PA14
Muralidhar Tata, Fabian Amman, Vinay Pawar, Michael T. Wolfinger, Siegfried Weiss, Susanne Häussler, Udo Bläsi
Front. Microbiol. 8:2312 (2017)
doi:10.3389/fmicb.2017.02312 | PDF | Journal article
19. The SmAP1/2 Proteins of the Crenarchaeon Sulfolobus Solfataricus Interact with the Exosome and Stimulate A-Rich Tailing of Transcripts
Birgit Märtens, Linlin Hou, Fabian Amman, Michael T. Wolfinger, Elena Evguenieva-Hackenberg, Udo Bläsi
Nucleic Acids Res. 45: 7938–49 (2017)
doi:10.1093/nar/gkx437 | PDF | Journal article
18. NMR Structural Profiling of Transcriptional Intermediates Reveals Riboswitch Regulation by Metastable RNA Conformations
Christina Helmling, Anna Wacker, Michael T. Wolfinger, Ivo L. Hofacker, Martin Hengsbach, Boris Fürtig, Harald Schwalbe
J. Am. Chem. Soc. 139 (7):2647–56 (2017)
doi:10.1021/jacs.6b10429 | Journal article
2016
17. Cross-Regulation by CrcZ RNA Controls Anoxic Biofilm Formation in Pseudomonas Aeruginosa
Petra Pusic, Muralidhar Tata, Michael T. Wolfinger, Elisabeth Sonnleitner, Susanne Häussler, Udo Bläsi
Sci. Rep. 6 (39621) (2016)
doi:10.1038/srep39621 | PDF | Journal article
16. Transcriptome-Wide Effects of Inverted SINEs on Gene Expression and Their Impact on RNA Polymerase II Activity
Mansoured Tajadodd, Andrea Tanzer, Konstantin Licht, Michael T. Wolfinger, Stefan Badelt, Florian Huber, Oliver Pusch, Sandy Schopoff, Ivo L. Hofacker, Michael F. Jantsch
Genome Biol. 17:220 (2016)
doi:10.1186/s13059-016-1083-0 | PDF | Journal article
15. Differential Transcriptional Responses to Ebola and Marburg Virus Infection in Bat and Human Cells
Martin Hölzer, Verena Krähling, Fabian Amman, Emanuel Barth, Stephan H. Bernhart, Victor Carmelo, Maximilian Collatz, Gero Doose, Florian Eggenhofer, Jan Ewald, Jörg Fallmann, Lasse M. Feldhahn, Markus Fricke, Juliane Gebauer, Andreas J. Gruber, Franziska Hufsky, Henrike Indrischek, Sabina Kanton, Jörg Linde, Nelly Mostajo, Roman Ochsenreiter, Konstantin Riege, Lorena Rivarola-Duarte, Abdullah H. Sahyoun, Sita J. Saunders, Stefan E. Seemann, Andrea Tanzer, Bertram Vogel, Stefanie Wehner, Michael T. Wolfinger, Rolf Backofen, Jan Gorodkin, Ivo Grosse, Ivo L. Hofacker, Steve Hoffmann, Christoph Kaleta, Peter F. Stadler, Stephan Becker, Manja Marz
Sci. Rep. 6 (34589) (2016)
doi:10.1038/srep34589 | PDF | Journal article
14. The MazF-Regulon: A Toolbox for the Post-Transcriptional Stress Response in Escherichia Coli
Martina Sauert, Michael T. Wolfinger, Oliver Vesper, Christian Müller, Konstantin Byrgazov, Isabella Moll
Nucleic Acids Res. 44 (14):6660–75 (2016)
doi:10.1093/nar/gkw115 | PDF | Journal article
13. Predicting RNA Structures from Sequence and Probing Data
Ronny Lorenz, Michael T. Wolfinger, Andrea Tanzer, Ivo L. Hofacker
Methods 103:86–98 (2016)
doi:10.1016/j.ymeth.2016.04.004 | PDF | Review article
12. RNA-Seq Based Transcriptional Profiling of Pseudomonas Aeruginosa Pa14 After Short- and Long-Term Anoxic Cultivation in Synthetic Cystic Fibrosis Sputum Medium
Muralidhar Tata, Michael T. Wolfinger, Fabian Amman, Nicole Roschanski, Andreas Dötsch, Elisabeth Sonnleitner, Susanne Häussler, Udo Bläsi
PLoS ONE 11 (1): e0147811 (2016)
doi:10.1371/journal.pone.0147811 | PDF | Journal article
11. SHAPE Directed RNA Folding
Ronny Lorenz, Dominik Luntzer, Ivo L. Hofacker, Peter F. Stadler, Michael T. Wolfinger
Bioinformatics 32: 145–47 (2016)
doi:10.1093/bioinformatics/btv523 | PDF | Journal article
2015
10. General and miRNA-Mediated mRNA Degradation Occurs on Ribosome Complexes in Drosophila Cells
Sanja Antic, Michael T. Wolfinger, Anna Skucha, Stefanie Hosiner, Silke Dorner
Mol. Cell. Biol. MCB–01346 (2015)
doi:10.1128/MCB.01346-14 | PDF | Journal article
9. ViennaNGS: A Toolbox for Building Efficient Next-Generation Sequencing Analysis Pipelines
Michael T. Wolfinger, Jörg Fallmann, Florian Eggenhofer, Fabian Amman
F1000Research 4:50 (2015)
doi:10.12688/f1000research.6157.2 | PDF | Journal article
2014
8. Memory Efficient RNA Energy Landscape Exploration
Martin Mann, Marcel Kucharík, Christoph Flamm, Michael T. Wolfinger
Bioinformatics 30: 2584–91 (2014)
doi:10.1093/bioinformatics/btu337 | PDF | Journal article
7. TSSAR: TSS Annotation Regime for dRNA-Seq Data
Fabian Amman, Michael T. Wolfinger, Ronny. Lorenz, Ivo L. Hofacker, Peter F. Stadler, Sven Findeiß
BMC Bioinformatics 15 (1) (2014)
doi:10.1186/1471-2105-15-89 | PDF | Journal article
2010
6. BarMap: RNA Folding on Dynamic Energy Landscapes
Ivo L. Hofacker, Christoph Flamm, Michael Heine, Michael T. Wolfinger, Gerik Scheuermann, Peter F. Stadler
RNA 16:1308–16 (2010)
doi:10.1261/rna.2093310 | PDF | Journal article
2008
5. Folding Kinetics of Large RNAs
Michael Geis, Christoph Flamm, Michael T. Wolfinger, Andrea Tanzer, Ivo L. Hofacker, Martin Middendorf, Christian Mandl, Peter F. Stadler, Caroline Thurner
J. Mol. Biol. 379 (1): 160–73 (2008)
doi:10.1016/j.jmb.2008.02.064 | Preprint PDF | Journal article
2006
4. Exploring the Lower Part of Discrete Polymer Model Energy Landscapes
Michael T. Wolfinger, Sebastian Will, Ivo L. Hofacker, Rolf Backofen, Peter F. Stadler
Europhys. Lett. 74(4): 726–32 (2006)
doi:10.1209/epl/i2005-10577-0 | Preprint PDF | Journal article
3. Visualization of Lattice-Based Protein Folding Simulations
Sebastian Pötzsch, Gerik Scheuermann, Peter F. Stadler, Michael T. Wolfinger, Christoph Flamm
In IV '06 Proceedings of the Conference on Information Visualization, pp89–94. Washington, DC, USA: IEEE Computer Society (2006)
doi:10.1109/IV.2006.127 | Conference article
2004
2. Efficient Computation of RNA Folding Dynamics
Michael T. Wolfinger, W. Andreas Svrcek-Seiler, Christoph Flamm, Ivo L. Hofacker, Peter F. Stadler
J. Phys. A: Math. Gen. 37(17): 4731–41 (2004)
doi:10.1088/0305-4470/37/17/005 | PDF | Journal article
2002
1. Barrier Trees of Degenerate Landscapes
Christoph Flamm, Ivo L. Hofacker, Peter F. Stadler, Michael T. Wolfinger
Z. Phys. Chem. 216: 155–73 (2002)
doi:10.1524/zpch.2002.216.2.155 | Preprint PDF | Journal article
- - -