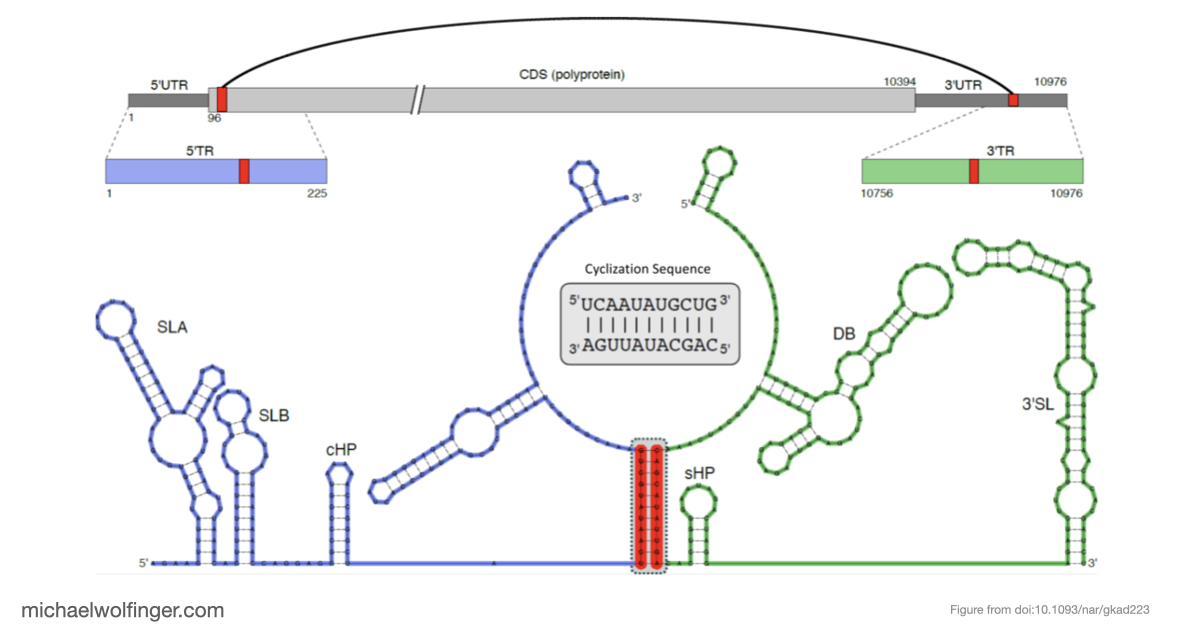
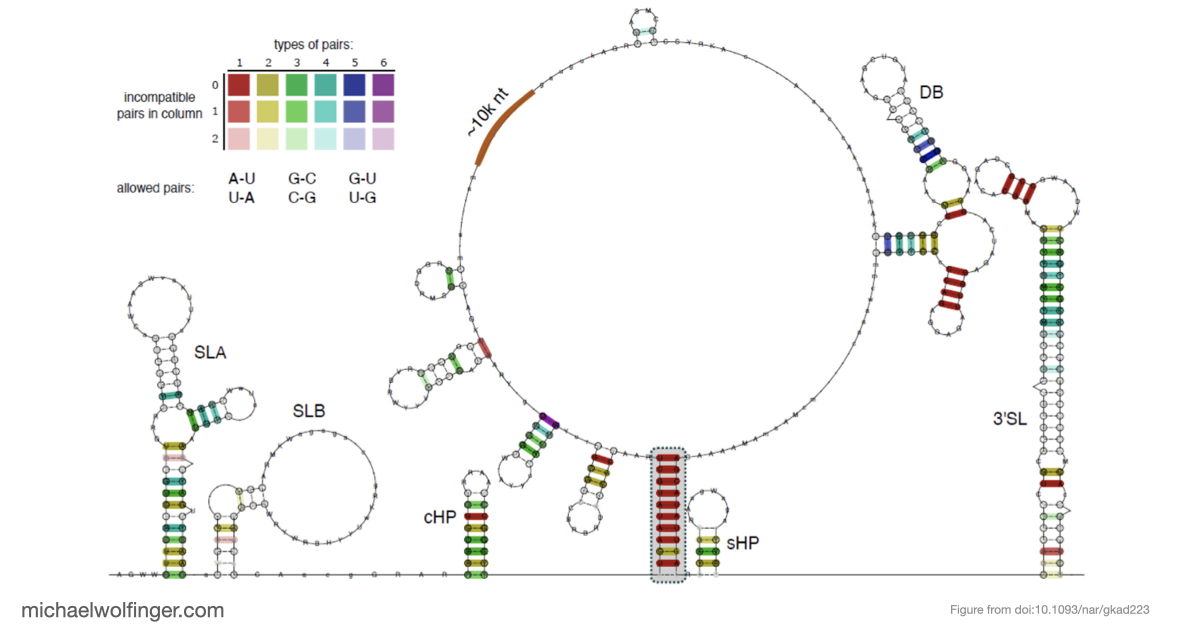
RNA–RNA interaction analysis of Japanese encephalitis virus
Unraveling the intricate RNA-RNA interactions in Japanese encephalitis virus (JEV) opens new possibilities for understanding viral replication and designing antiviral therapeutics, as our recent study reveals the importance of a conserved cyclization sequence in driving these interactions
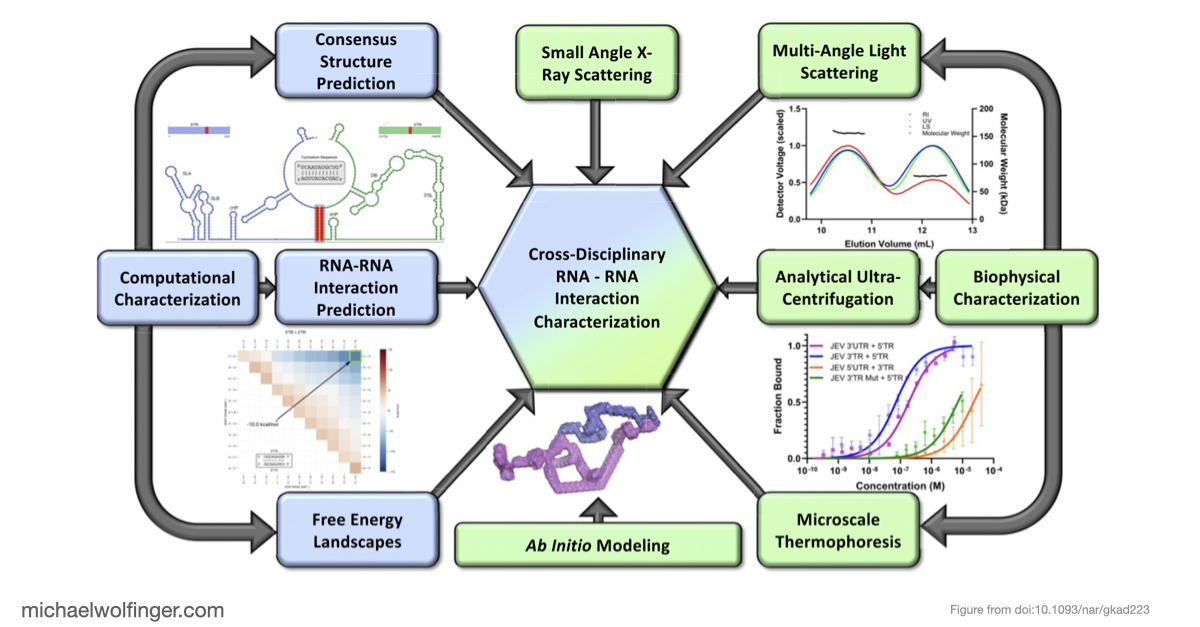
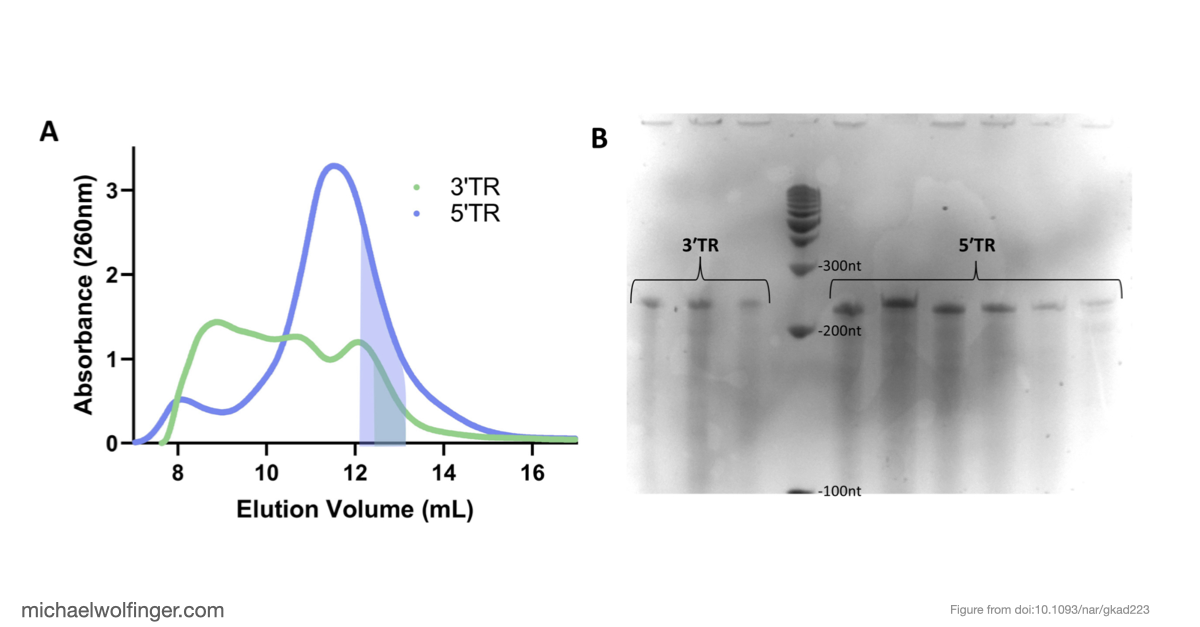
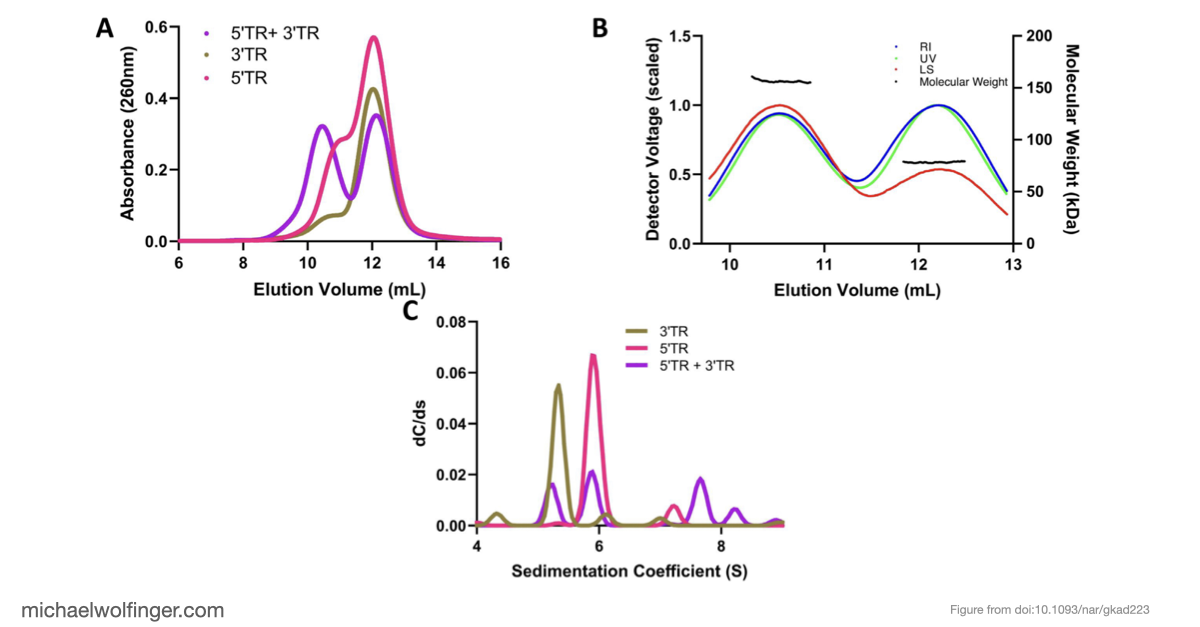
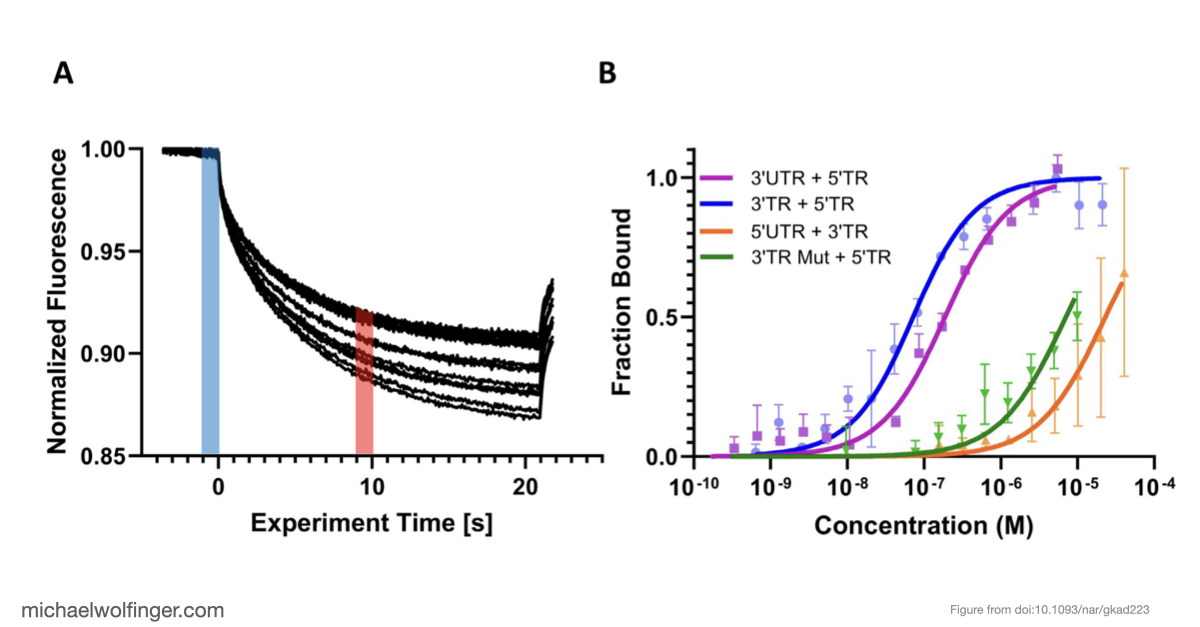
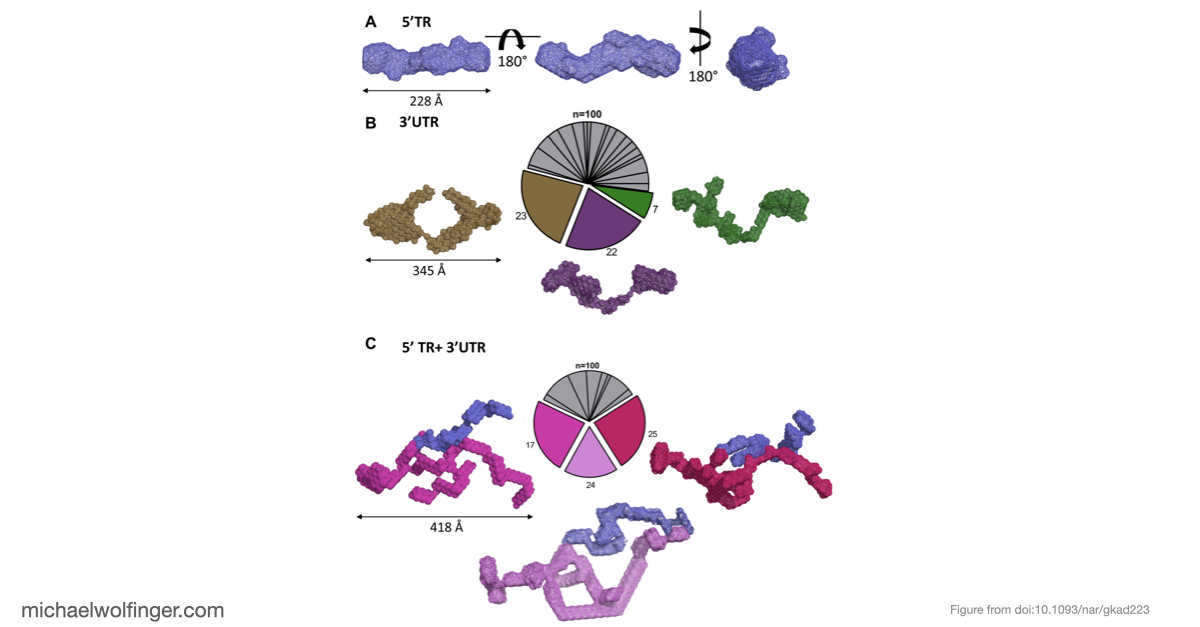
In this study, we have utilized a unique combination of computational and biophysical techniques to investigate RNA-RNA interactions in Japanese encephalitis virus (JEV). We identified a specific JEV isolate with a hypothesized high-affinity interaction, and then employed various biophysical characterization methods to validate and characterize this interaction. Through size-exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) and analytical ultracentrifugation (AUC), we demonstrated the direct interaction between the 5' and 3' terminal regions (TRs) of JEV with nanomolar affinity and 1:1 stoichiometry.
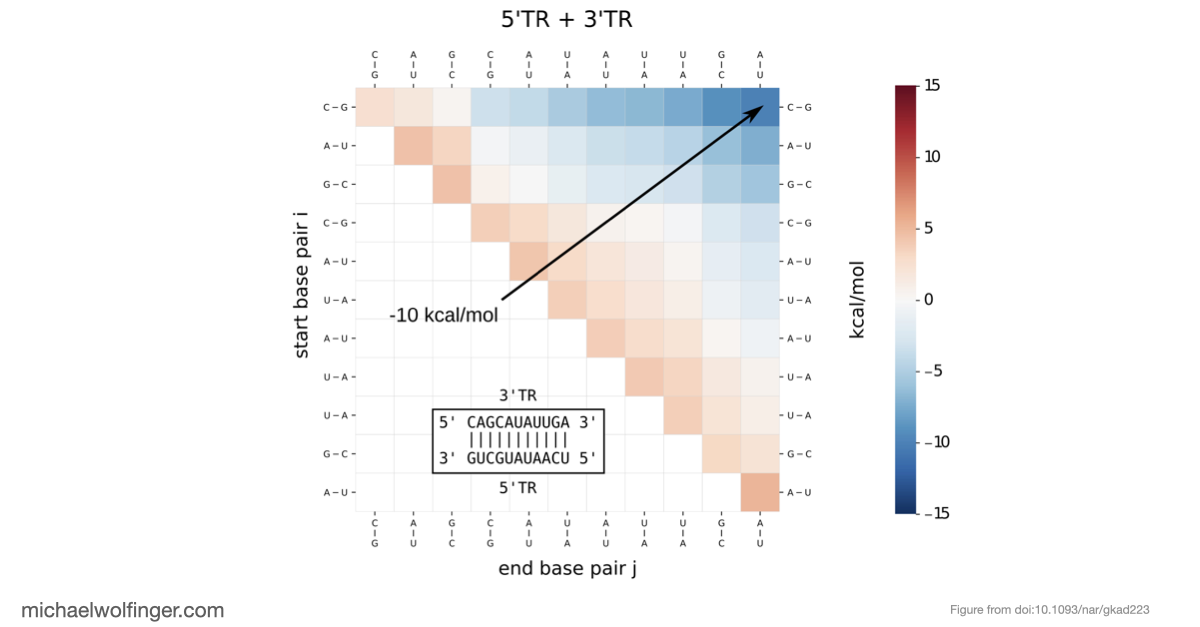
Our computational analysis supported a kinetic favorability of the cyclization sequence interaction and its competitive advantage over potential homo-dimer RNA interactions. This provides valuable insights into the replication pathway/mechanism of flavivirus NS5 in replicating the viral genome. Our study also presents a low-resolution ab initio model that offers a potential architectural arrangement of this RNA-RNA interaction in solution. These findings lay a solid foundation for the development of pharmaceutical therapies targeting viral replication through interruption of the cyclization sequence, and also shed light on the fundamental understanding of RNA-RNA interactions in flaviviruses and other viral systems.
The study's insights into RNA-RNA interactions in Japanese encephalitis virus align with the One Health approach, highlighting the potential for antiviral therapies to benefit both human and animal health while considering the broader ecosystem.
Figures and Data
Citation
Investigating RNA-RNA interactions through computational and biophysical analysis
Tyler Mrozowich, Sean Park, Maria Waldl, Amy Henrickson, Scott Tersteeg, Corey R. Nelson, Anneke De Klerk, Borries Demeler, Ivo L. Hofacker, Michael T. Wolfinger, Trushar R. Patel
Nucleic Acids Res. gkad223 (2023) | doi:10.1093/nar/gkad223 | PDF | Supplement