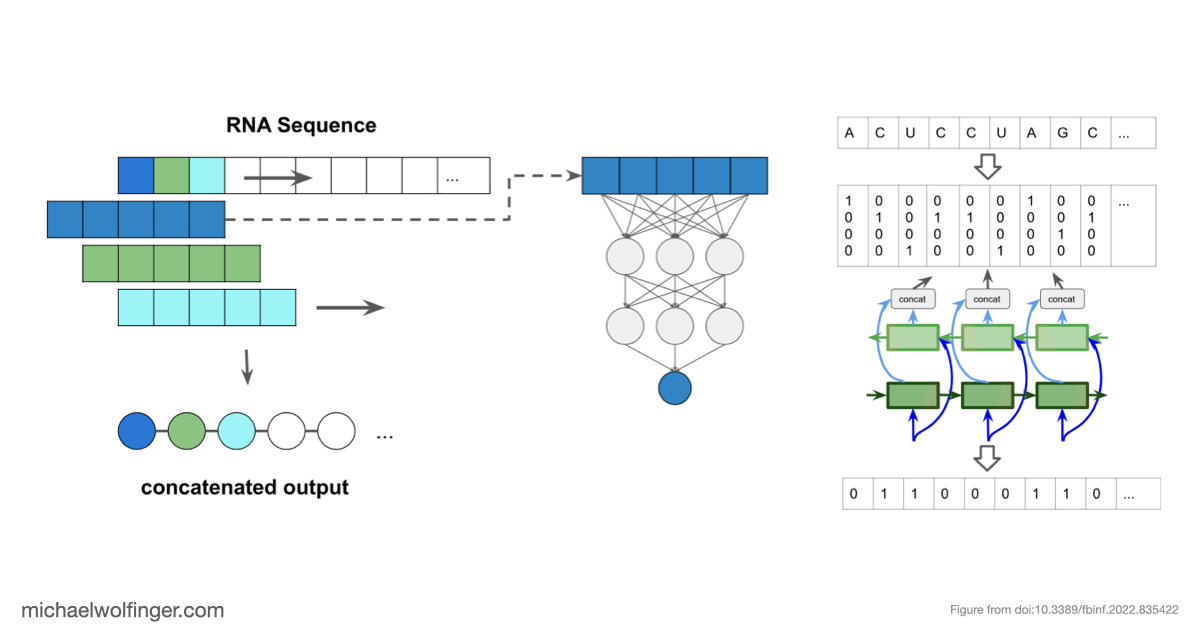
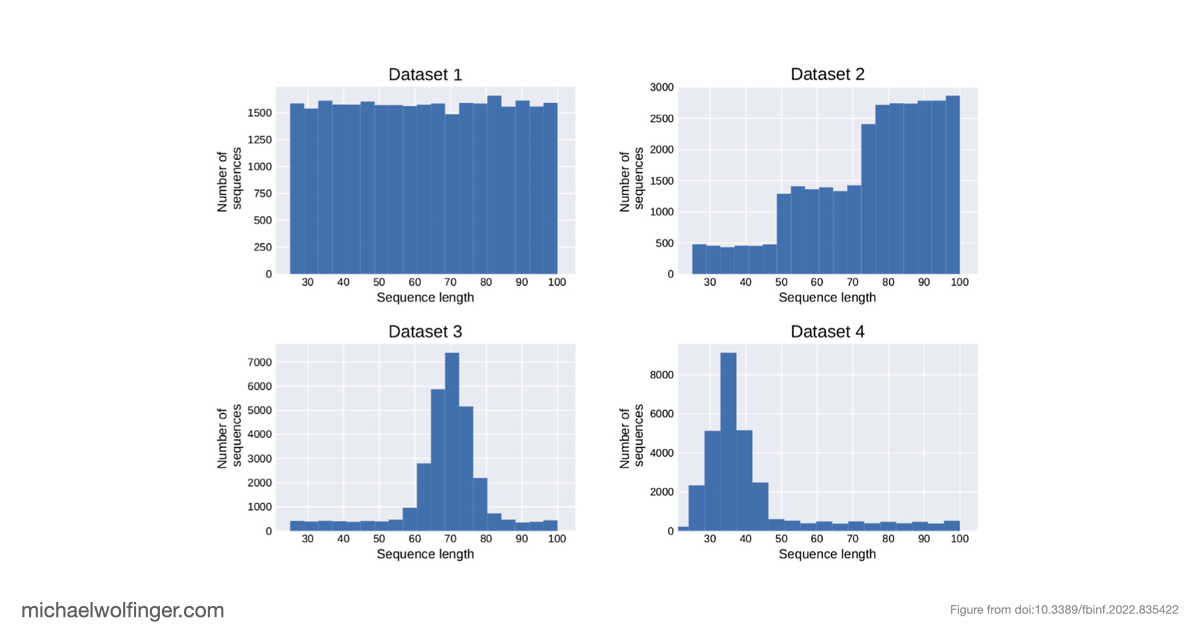
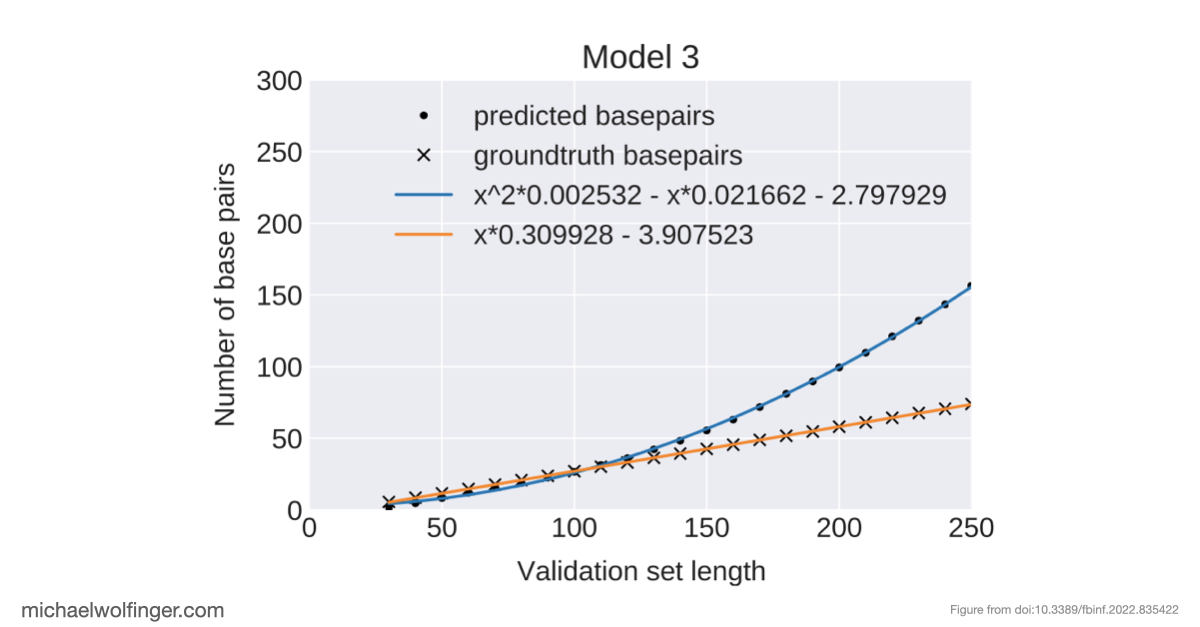
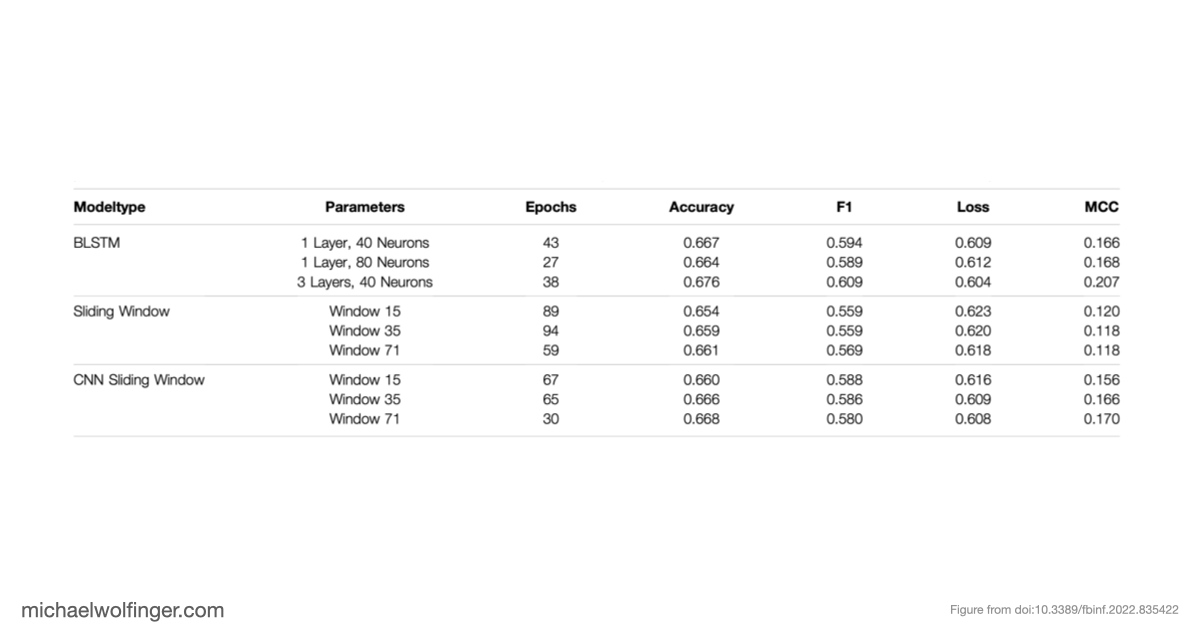
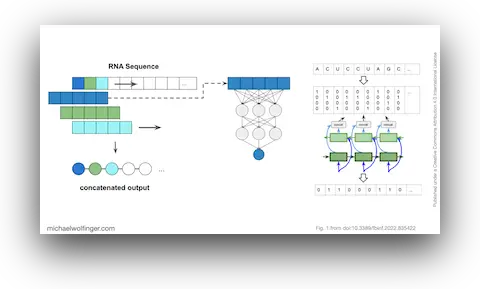
Deep learning methods are unable to predict RNA secondary structures
RNA structure prediction might seem like an ideal fit for machine learning, but it's more challenging than you might think. In this paper, we explore these difficulties by using synthetic data generated by ViennaRNA's RNAfold, offering a controlled environment to study how neural networks handle RNA secondary structure prediction. What we found suggests that the limitations seen in artificial settings can directly translate to real-world data, raising important questions about the effectiveness of current machine learning approaches in this field.
One of the major issues we discovered is that many published AI models for RNA structure prediction rely on highly biased training sets. This bias significantly impacts the accuracy of predictions when the models are tested on RNA sequences outside of their training set. By using a technique known as inverse RNA folding—where we generate sequences that match specific structures—we created synthetic data with the same kind of bias found in these deep learning models. When we trained a neural network on this biased data, it performed well on new sequences that folded into familiar structures. However, when faced with sequences forming novel structures, the performance dropped drastically. This shows that while the network could generalize to new sequences, it struggled to predict new or unseen structures.
Even more surprising is that many deep learning methods trained on unbiased data couldn't even reliably predict base pairing—a fundamental aspect of RNA structure that's simpler than the full RNA folding problem. Additionally, certain models, such as BLSTMs, predicted structural features like pseudoknots and base triplets, which don’t even appear in the ViennaRNA RNAfold ground truth. This mismatch between model output and reality underscores the limitations of these approaches.
Our findings reveal significant gaps in current machine learning models for RNA structure prediction and highlight the need for more robust training methods. These insights could drive future improvements in how AI is applied to RNA biology, especially for tackling more complex tasks like RNA folding.
Figures and Data
Citation
Caveats to deep learning approaches to RNA secondary structure prediction
Christoph Flamm, Julia Wielach, Michael T. Wolfinger, Stefan Badelt, Ronny Lorenz, Ivo L. Hofacker
Front. Bioinform. 2:835422 (2022) | doi:10.3389/fbinf.2022.835422 | PDF