BVDV IRES RNA in focus: A game changer for livestock health
Here we characterize a pseudoknot in the 5'UTR Internal Ribosome Entry Site (IRES) of Bovine Viral Diarrhea Virus (BVDV) using SHAPE-guided RNA structure probing and computational modelling of secondary and tertiary structures
RNA viruses have long fascinated researchers with their ability to adapt and evolve using minimal genetic material. Among these viruses, Bovine Viral Diarrhea Virus (BVDV) stands out as a significant veterinary pathogen, causing substantial economic losses in the livestock industry worldwide. Understanding the molecular mechanisms behind the replication cycle of BVDV is crucial for developing effective strategies to combat this virus. In our recently published paper, we delve into the intricacies of BVDV's RNA structure, shedding light on its critical role in viral translation. Here, we aim to provide an accessible explanation of our findings for both experts and non-virologists and emphasize the relevance of our research to the One Health System.
BVDV belongs to the Pestivirus genus, a group of viruses known for their economic impact on the livestock industry. Like other RNA viruses, BVDV employs resource-efficient molecular strategies to optimize its replication cycle. Central to this process is the Internal Ribosome Entry Site (IRES) located within the virus's 5' Untranslated Region (UTR). The IRES is a complex structural domain that controls cap-independent translation, a crucial step in viral infection.
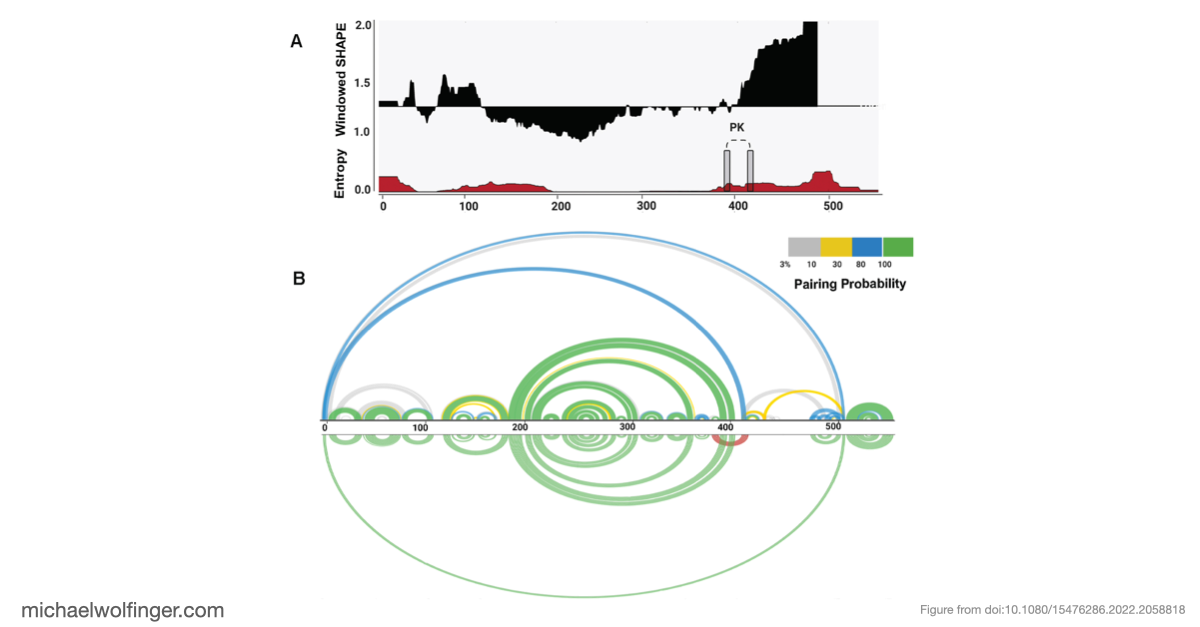
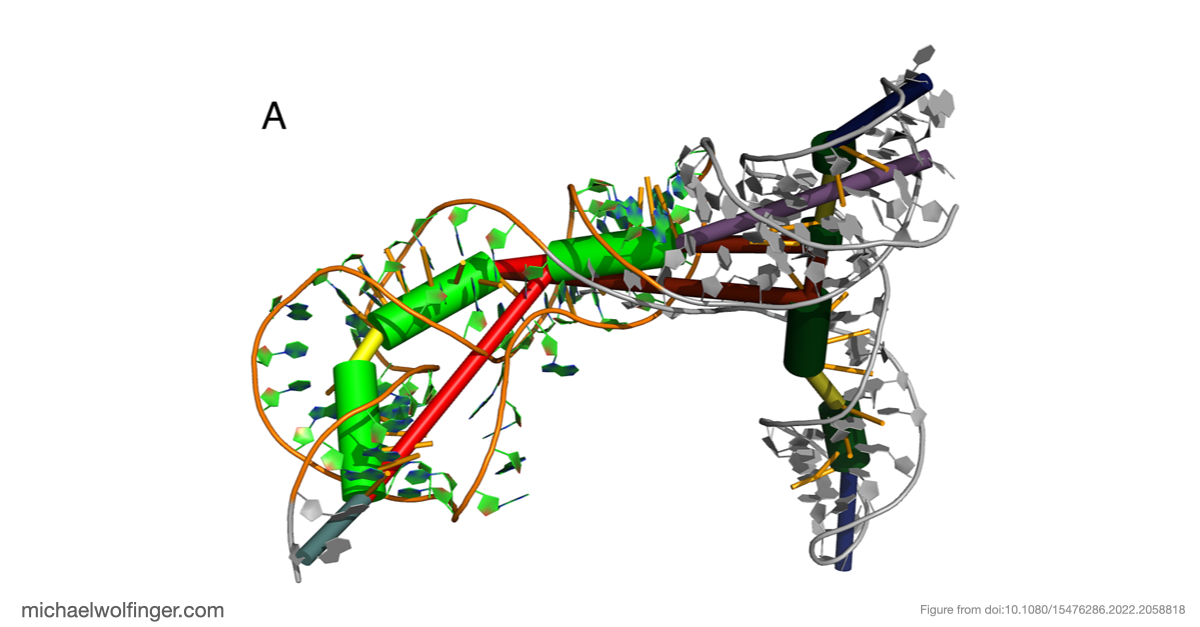
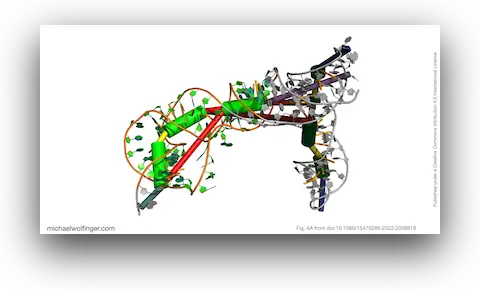
Our study focused on unraveling the intricate secondary and tertiary structure of BVDV IRES RNA. We used RNA structural probing methods and computational 3D modeling to gain insights into the structural arrangement of this critical RNA component.
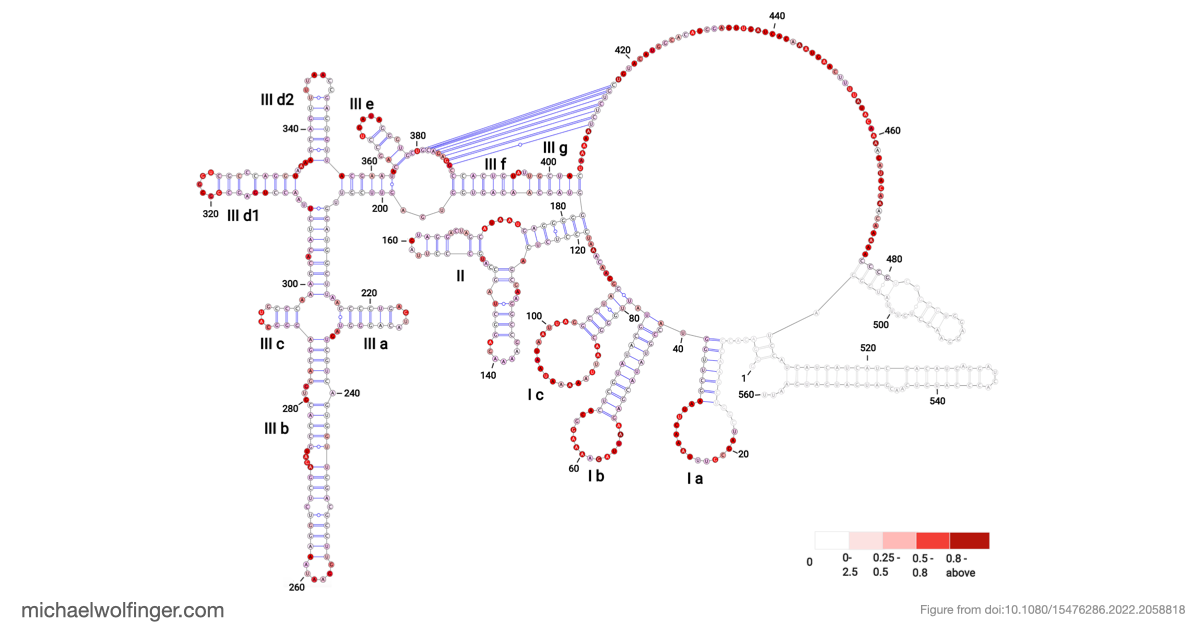
Our research revealed that BVDV IRES RNA consists of three major structural domains (I-III), each containing distinct motifs, such as hairpins and stems, interspersed among less structurally constrained regions. While our model aligned with previous secondary structure models for BVDV strains, we identified an alternative folding pattern in domain II. This structural flexibility in domain II was supported by thermodynamic parameters, highlighting the dynamic nature of BVDV IRES RNA.
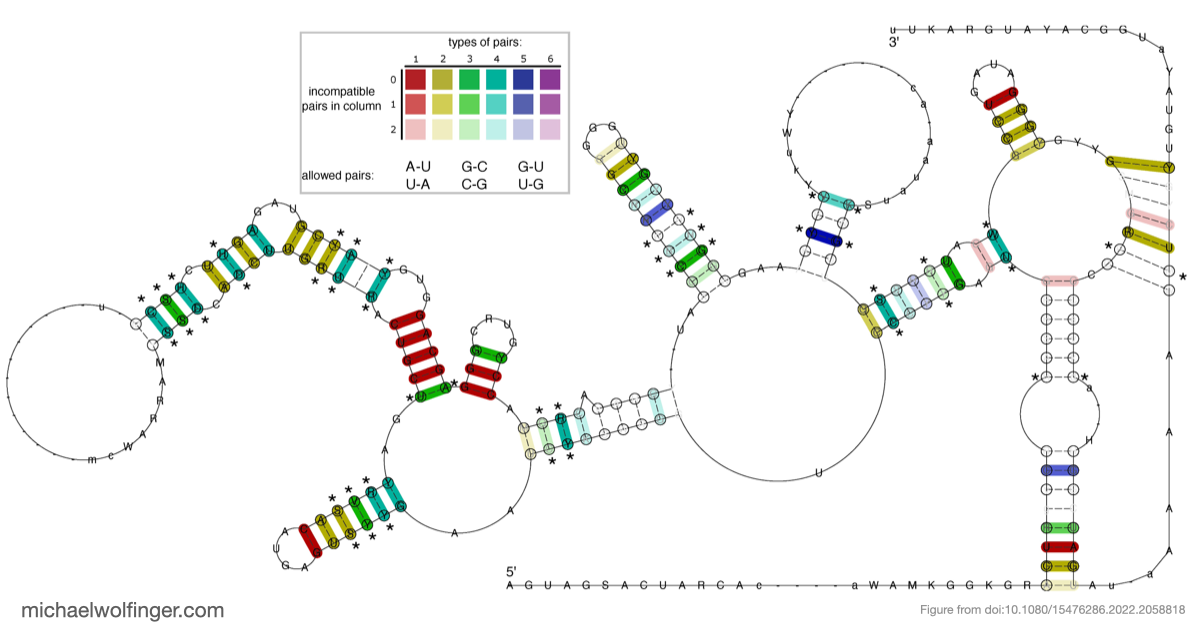
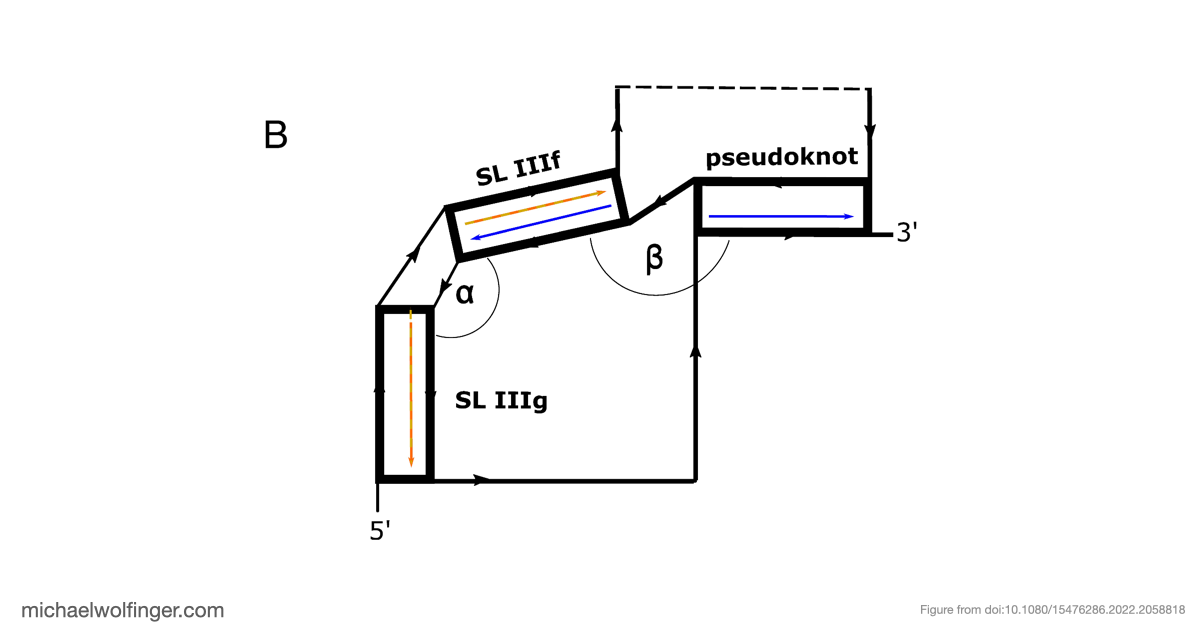
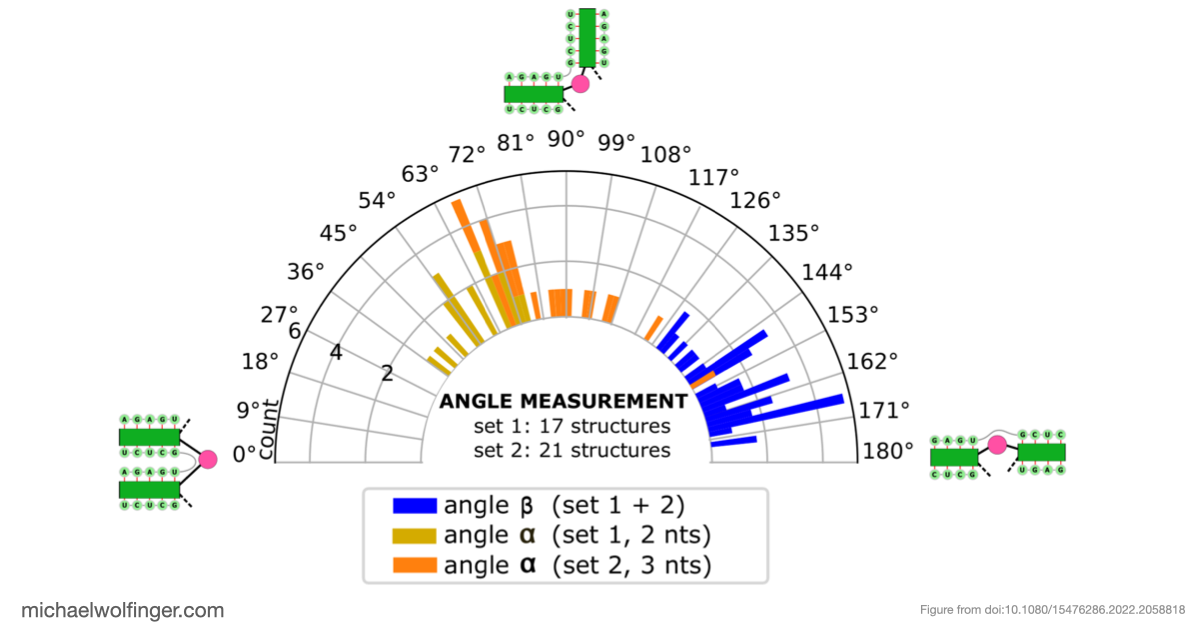
The most significant discovery in our study pertains to domain III, which exhibited high structural stability. It includes single-stranded regions and a pseudoknot, a structural element crucial for viral translation. Our 3D modeling revealed quasi-coaxial stacking between specific motifs in domain III, providing insight into the stability of the IRES pseudoknot.
Previous studies had suggested the presence of a pseudoknot in BVDV IRES RNA, but our research provided computational evidence supporting its crucial role. We conducted a comprehensive analysis of BVDV IRES RNA sequences from different genotypes and identified statistically significant covariation in 22 base pairs, including the pseudoknot.
Our findings have far-reaching implications in combating BVDV and related viruses. The similarities between BVDV and Hepatitis C Virus (HCV) IRES RNAs suggest that strategies developed to target HCV IRES RNA could potentially be applied to interfere with BVDV replication. This includes the identification of small molecule inhibitors, antisense oligonucleotides, and ribozymes to disrupt the IRES RNA, preventing the binding of translational factors and ultimately inhibiting viral replication.
In conclusion, our research has unveiled the structural intricacies of BVDV IRES RNA, shedding light on its role in viral translation. This knowledge opens up exciting possibilities for the development of targeted therapies against BVDV and related viruses, contributing to the One Health System's mission of protecting both human and animal health. As we continue to delve deeper into the molecular mechanisms of these pathogens, we move one step closer to mitigating the threats they pose to our global community.
Figures and Data
Citation
Insights into the secondary and tertiary structure of the Bovine Viral Diarrhea Virus Internal Ribosome Entry Site
Devadatta Gosavi, Iwona Wower, Irene K Beckmann, Ivo L Hofacker, Jacek Wower, Michael T Wolfinger, Joanna Sztuba-Solinska
RNA Biol. 19(1) 496-506 (2022) | doi:10.1080/15476286.2022.2058818 | PDF