Molecular epidemiology and RNA structures in Chikungunya virus
This research provides a comprehensive analysis of the molecular epidemiology and evolutionary dynamics of Chikungunya virus (CHIKV), uncovering how single nucleotide variants influence viral RNA structures and shape the global spread of the virus
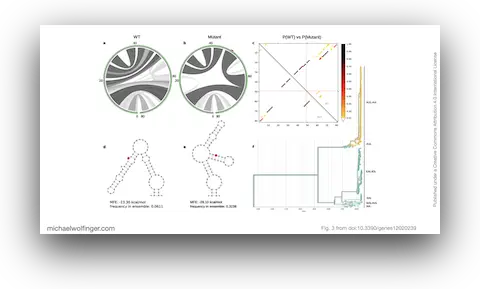
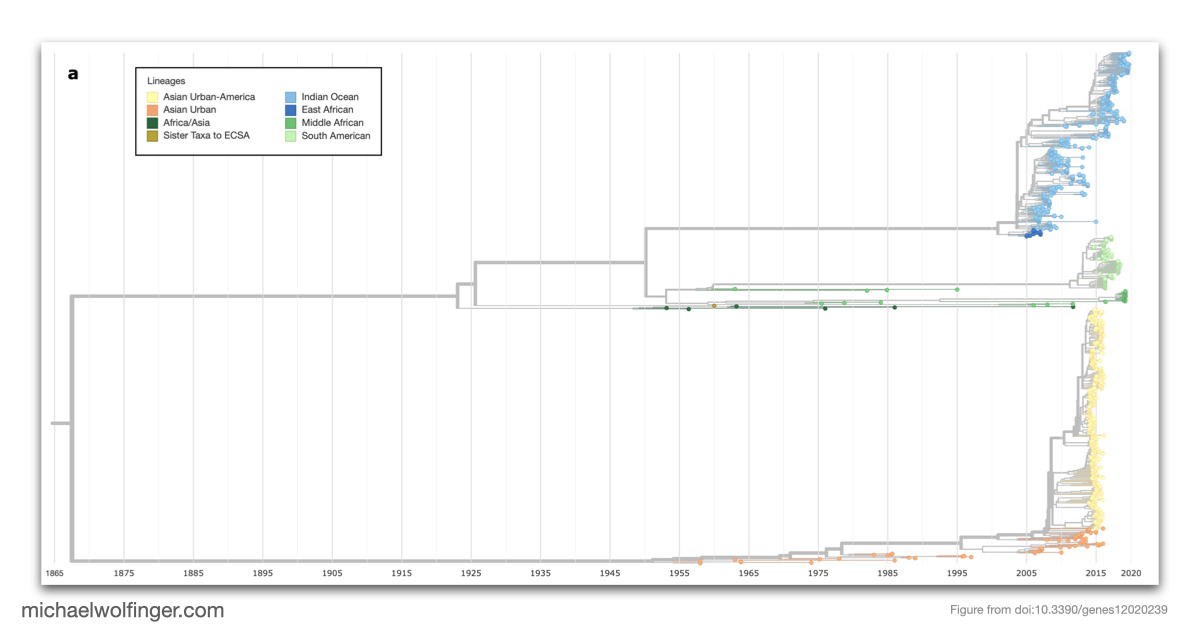
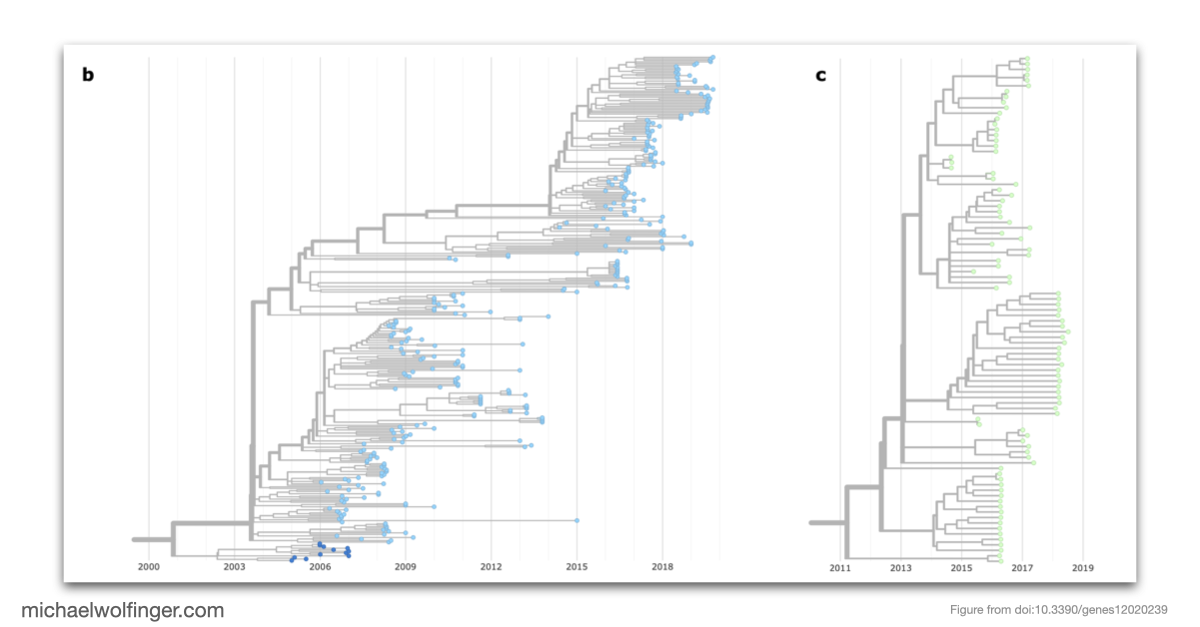
Chikungunya virus, an emerging Alphavirus, causes millions of human infections annually and has witnessed outbreaks in Africa and Asia since the 1950s. The research presented here delves into the molecular epidemiology of CHIKV, revealing how single nucleotide variants can impact the virus's RNA structures. Utilizing more than 1000 publicly available CHIKV genomes, the study provides an interactive phylodynamics dataset using Nextstrain, a tool for real-time tracking of pathogen evolution. The data set, publicly available through CHIKVnext, enables the tracking of spatiotemporal and epidemiological aspects of the CHIKV global spread.
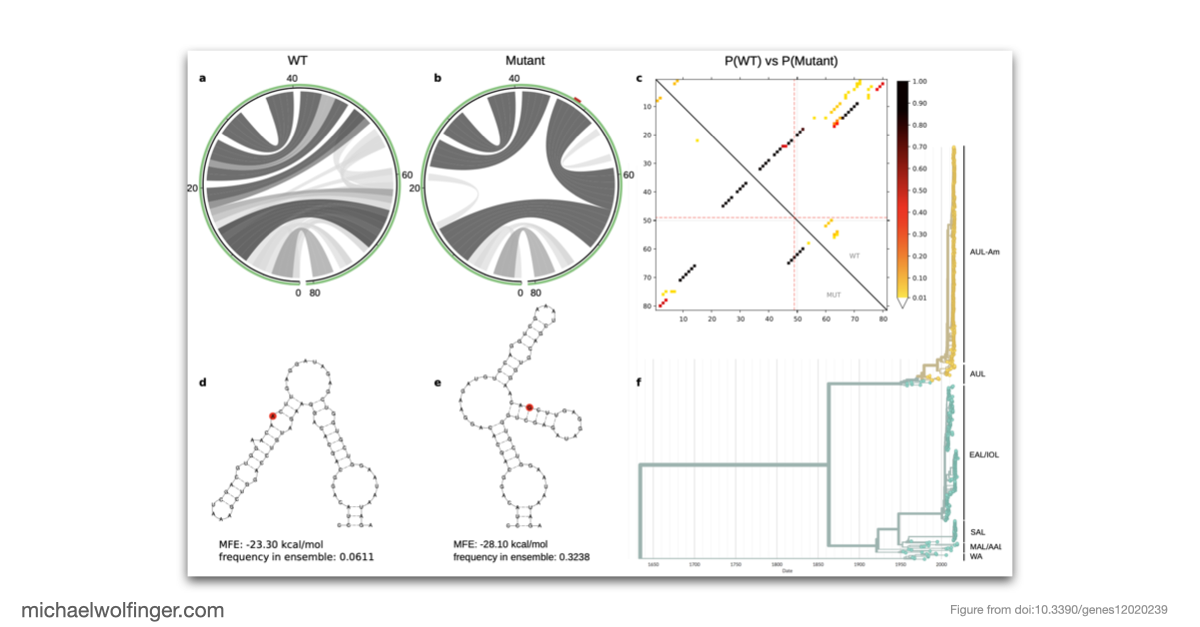
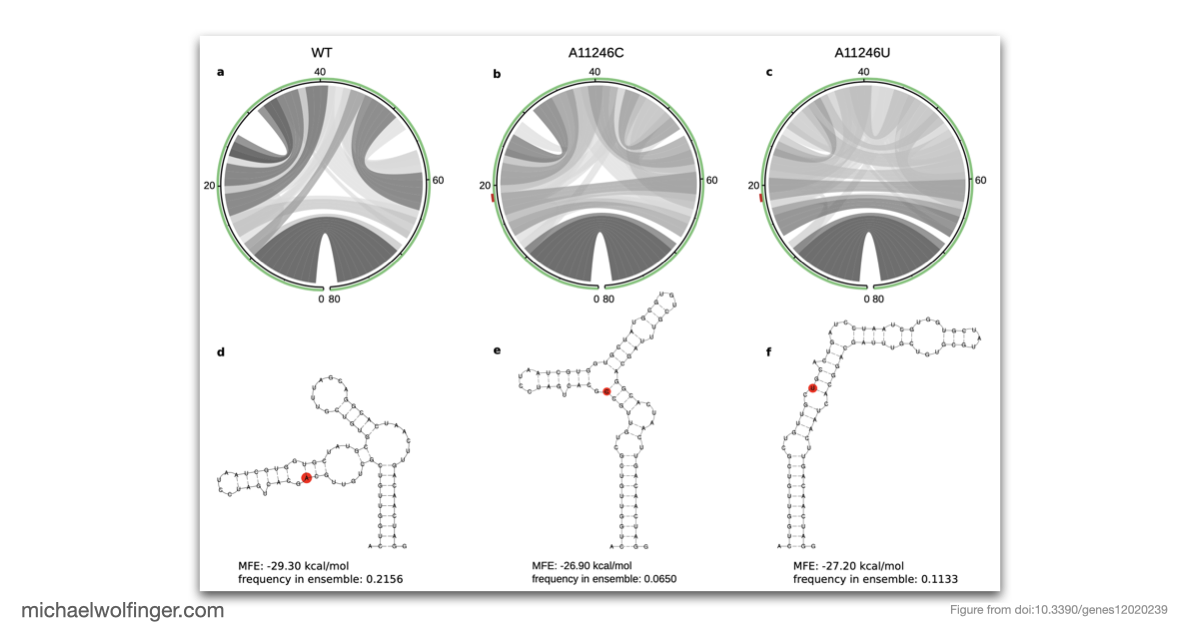
The study addresses genotype/phenotype associations in CHIKV by employing comparative approaches, molecular epidemiology concepts, phylogeny reconstruction, and computational RNA biology. The nucleotide divergence within CHIKV lineages is found to be relatively low, often due to geographical constraints and a limited collection period for newer lineages. However, the divergence between different lineages can be considerable, with the West African lineage showing the highest divergence compared to others.
This publication questions the current nomenclature of CHIKV lineages, and suggests that a new system, independent of geography, might be more coherent and beneficial for drug and vaccine development. Despite the low divergence in some established lineages, mutations like A226V in E1 can significantly impact the virus's capacity to replicate. The study further highlights the importance of increased surveillance to identify the virus at the time of introduction rather than during a pandemic.
This research exemplifies the importance of a holistic, interdisciplinary approach in understanding and combating infectious diseases such as CHIKV. The findings, which combine molecular epidemiology and RNA structure prediction, not only pave the way for better diagnostic and treatment methods but also underscore the necessity of a timely response to prevent outbreaks. The One Health approach is particularly pertinent here. By adopting a unified perspective, scientists, healthcare professionals, and policymakers can work together to address the multifaceted challenges posed by CHIKV and similar infectious diseases, ultimately contributing to the well-being of our global ecosystem.
Figures and Data
Citation
Dynamic Molecular Epidemiology Reveals Lineage-Associated Single-Nucleotide Variants That Alter RNA Structure in Chikungunya Virus
Thomas Spicher, Markus Delitz, Adriano de Bernardi Schneider, Michael T. Wolfinger
Genes 12 (2):239 (2021) | doi:10.3390/genes12020239 | PDF | Figures
See Also
Updated Phylogeny of Chikungunya Virus Suggests Lineage-Specific RNA Architecture
Adriano de Bernardi Schneider, Roman Ochsenreiter, Reilly Hostager, Ivo L. Hofacker, Daniel Janies, Michael T. Wolfinger
Viruses 11:798 (2019) | doi:10.3390/v11090798 | PDF | Figures