Mpulungu Virus: A glimpse into Africa's novel tick-borne flavivirus
The discovery of Mpulungu virus (MPFV), a novel tick-borne flavivirus in Zambia, expands our understanding of the geographical distribution and genetic diversity of flaviviruses across Africa. This study delves into the genetic details of MPFV, suggesting its potential to infect vertebrate hosts and underscoring the importance of a One Health approach in addressing emerging infectious diseases
We report the discovery of Mpulungu virus (MPFV), a novel tick flavivirus, identified in a tick species from Zambia. The complete genome sequence of MPFV was determined and found to be closely related to the Ngoye virus from Senegal, suggesting that this group of viruses may be widespread across Africa. This expands the known geographical distribution and genetic diversity of flaviviruses. Despite unsuccessful attempts to isolate MPFV in cell culture and neonatal mice, the study employed a predictive analysis based on dinucleotide ratios, which suggested that MPFV might infect vertebrate hosts similar to other tick-borne flaviviruses (TBFVs), potentially posing a public health risk.
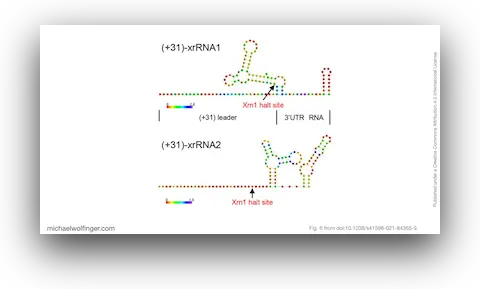
The study further delved into the genetic details of MPFV, focusing on the untranslated regions (UTRs) of the viral genome that are crucial for viral translation and replication. The 5′-UTR of MPFV showed a typical flavivirus structure, suggesting similar RNA synthesis processes. However, the 3′-UTR displayed some unique characteristics, differing from known TBFVs in terms of sequence composition, length, and secondary structures.
The study identified two exoribonuclease-resistant RNA (xrRNA) structures in the 3′-UTR of MPFV that achieve resistance to an enzyme called Xrn1, which is typically involved in degrading viral RNA. The presence of multiple xrRNA structures in MPFV's 3′-UTR was also noted to potentially contribute to the production of subgenomic flavivirus RNA (sfRNA). Past studies have suggested that xrRNA structures play a significant role in the viral life cycle and pathogenicity.
The discovery of MPFV in Zambia and its potential relation to Ngoye virus in Senegal points towards a need for comprehensive surveillance of flaviviruses across diverse ecosystems. By identifying and understanding the genetic makeup and host range of such viruses, preventive measures can be implemented to mitigate potential public health risks. This study highlights the necessity of integrated efforts and knowledge sharing among health professionals, veterinarians, and environmental scientists to enhance our preparedness and response to emerging infectious diseases. In light of these findings, employing the One Health approach becomes imperative to comprehensively address and understand the complexities and potential implications of emerging flaviviruses like MPFV.
Citation
An African Tick Flavivirus Forming an Independent Clade Exhibits Unique Exoribonuclease-Resistant RNA Structures in the Genomic 3’-Untranslated Region
Hayato Harima, Yasuko Orba, Shiho Torii, Yongjin Qiu, Masahiro Kajihara, Yoshiki Eto, Naoya Matsuta, Bernard M. Hang’ombe, Yuki Eshita, Kentaro Uemura, Keita Matsuno, Michihito Sasaki, Kentaro Yoshii, Ryo Nakao, William W. Hall, Ayato Takada, Takashi Abe, Michael T. Wolfinger, Martin Simuunza, Hirofumi Sawa
Sci. Rep. 11:4883 (2021) | doi: 10.1038/s41598-021-84365-9 | PDF
See Also
Xinyang flavivirus, from Haemaphysalis flava ticks in Henan province, China, defines a basal, likely tick-only flavivirus clade
Lan-Lan Wang, Qia Cheng, Natalee D. Newton, Michael T. Wolfinger, Mahali S. Morgan, Andrii Slonchak, Alexander A. Khromykh, Tian-Yin Cheng, Rhys H. Parry
J. Gen. Virol. 105(5) (2024) | doi:10.1099/jgv.0.001991 | PDF