Chikungunya Virus Phylogeny: Lineage-Specific RNA Structures
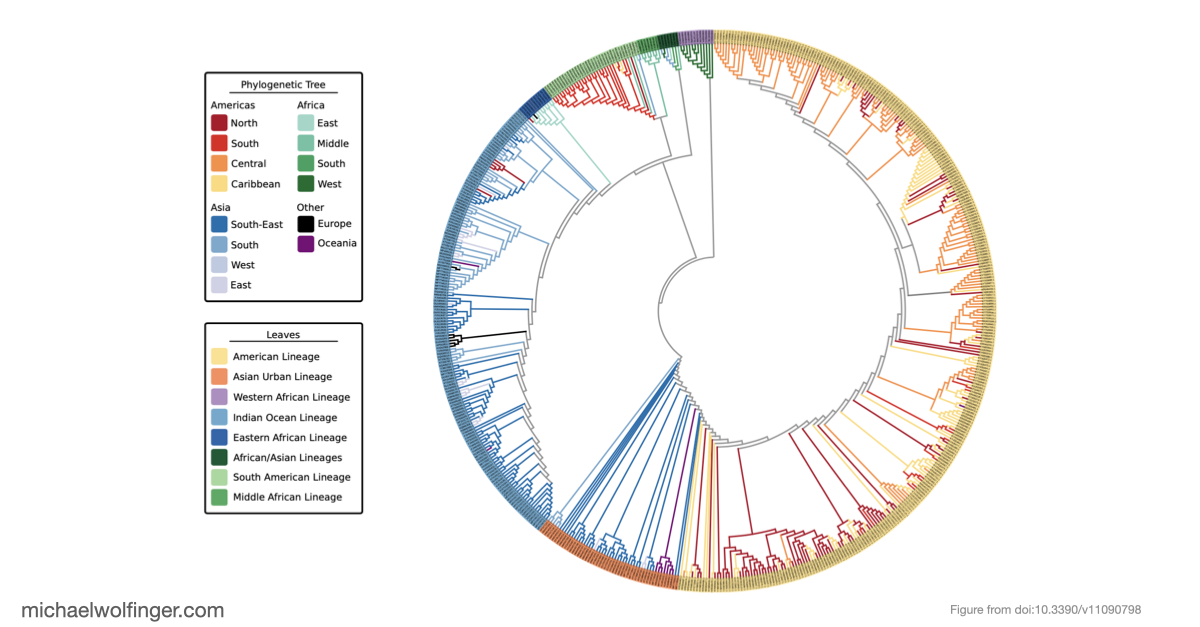
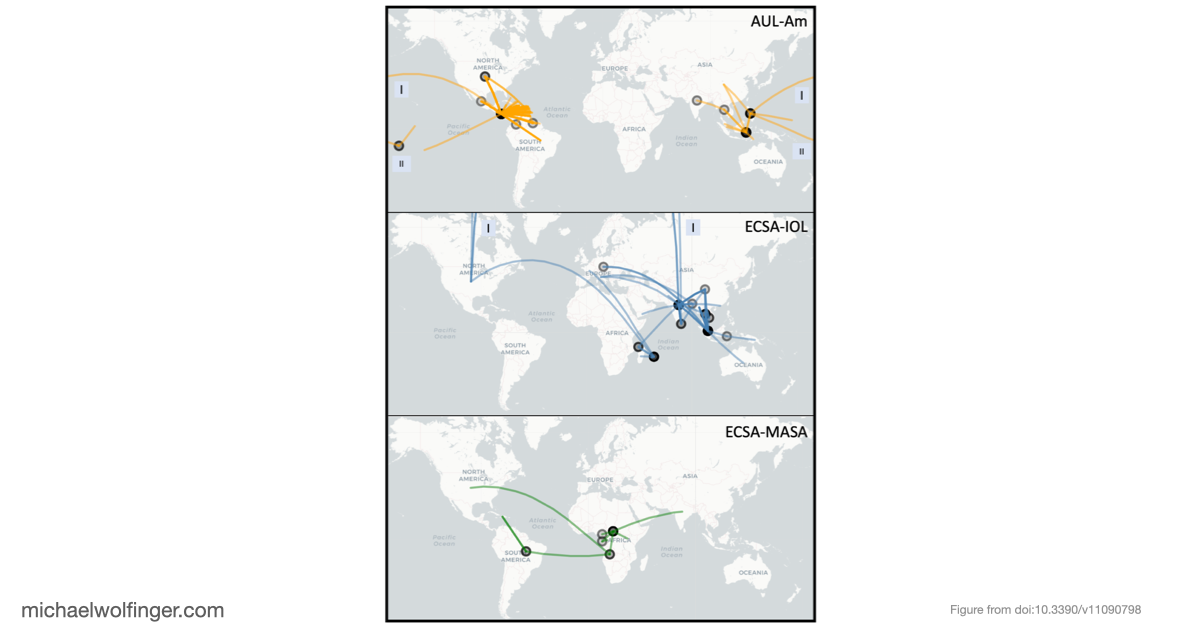
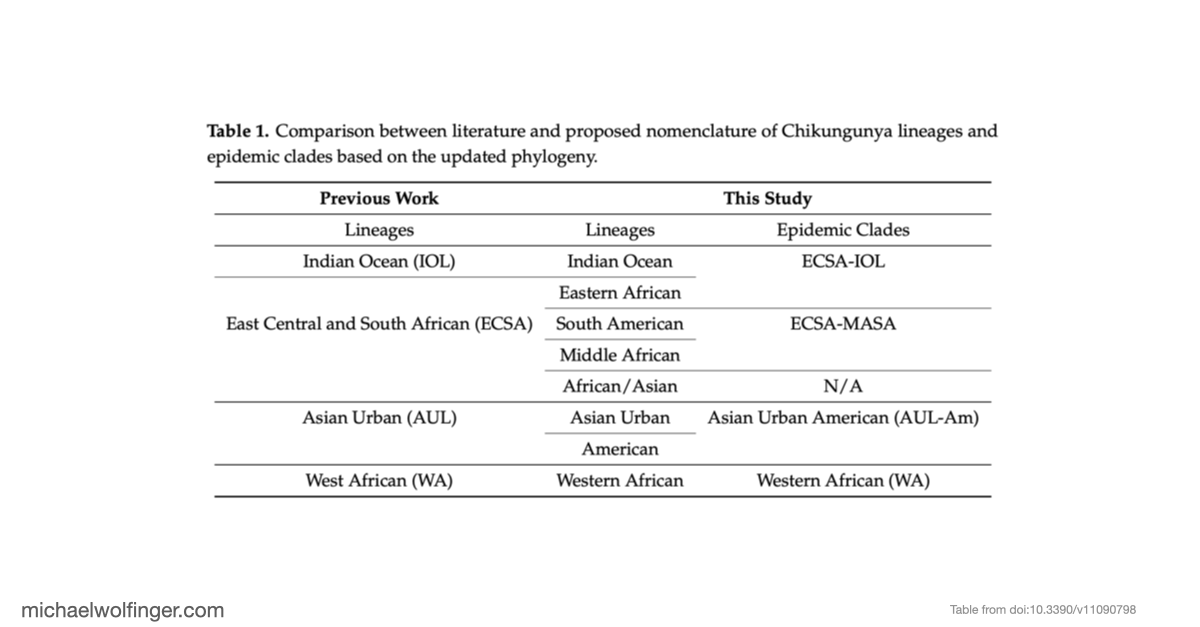
In this study, we analyzed the evolution and spread of the Chikungunya virus (CHIKV) across Africa by examining 598 whole genome sequences. Our analysis revealed several distinct CHIKV lineages with strong support, providing new insights into the virus's evolutionary history and its movement across regions
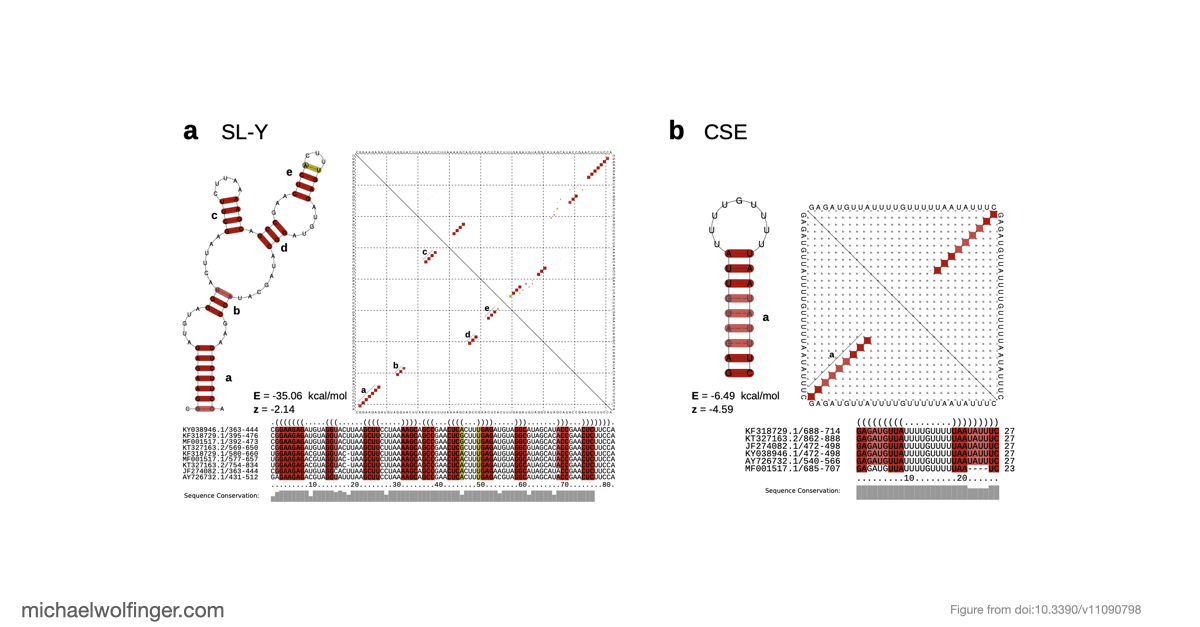
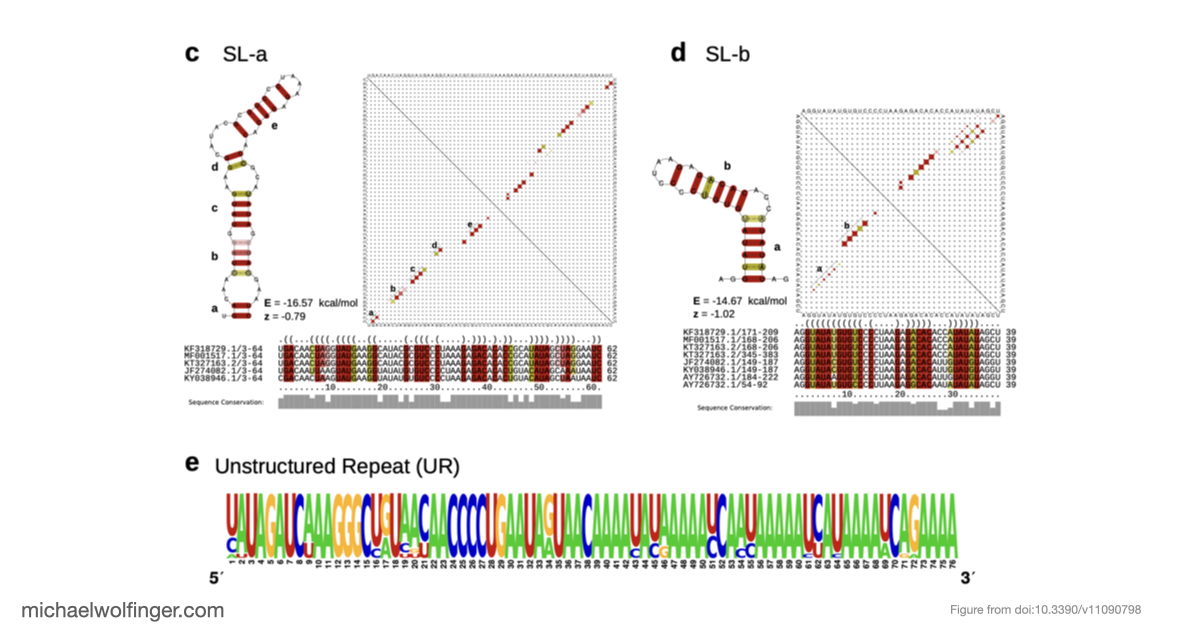
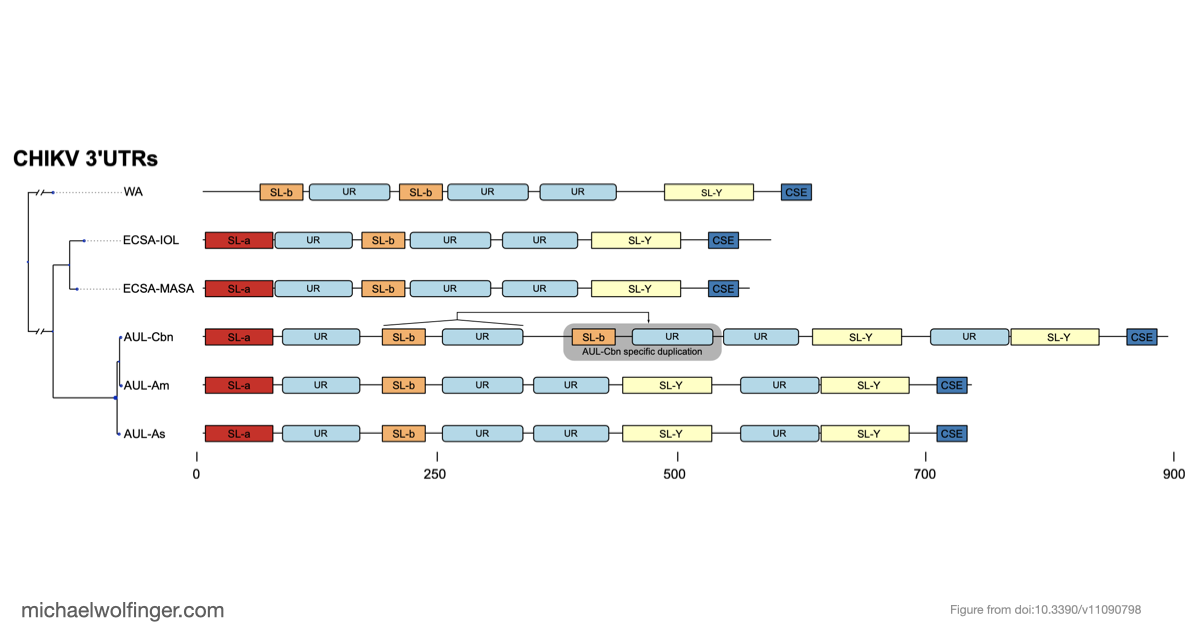
We carried out an in-depth analysis of the virus’s 3’UTR, where we uncovered conserved RNA structures that likely play crucial roles in stabilizing viral RNA and interacting with host proteins. One of our most intriguing findings was the discovery of unique 3’UTR structures specific to certain lineages. These features may help the virus counter host defenses and interact with key cellular proteins, such as the Musashi family of translational regulators.
Our comprehensive analysis provides valuable insights into the complex evolution and epidemiology of CHIKV. By highlighting lineage-specific traits, we hope this study will pave the way for a deeper understanding of viral genomics and open up new opportunities for targeted interventions. As we continue to explore the intricacies of CHIKV's genetic makeup, our findings represent an important step in the global effort to combat this significant public health threat.
Figures and Data
Citation
Updated Phylogeny of Chikungunya Virus Suggests Lineage-Specific RNA Architecture
Adriano de Bernardi Schneider, Roman Ochsenreiter, Reilly Hostager, Ivo L. Hofacker, Daniel Janies, Michael T. Wolfinger
Viruses 11:798 (2019) | doi:10.3390/v11090798 | PDF | Figures
See Also
Dynamic Molecular Epidemiology Reveals Lineage-Associated Single-Nucleotide Variants That Alter RNA Structure in Chikungunya Virus
Thomas Spicher, Markus Delitz, Adriano de Bernardi Schneider, Michael T. Wolfinger
Genes 12 (2):239 (2021) | doi:10.3390/genes12020239 | PDF | Figures